| bacterial_genetics_10_10_2018combo.pptx | |
| File Size: | 31970 kb |
| File Type: | pptx |
|
•Functional unit of genetic information is the gene.
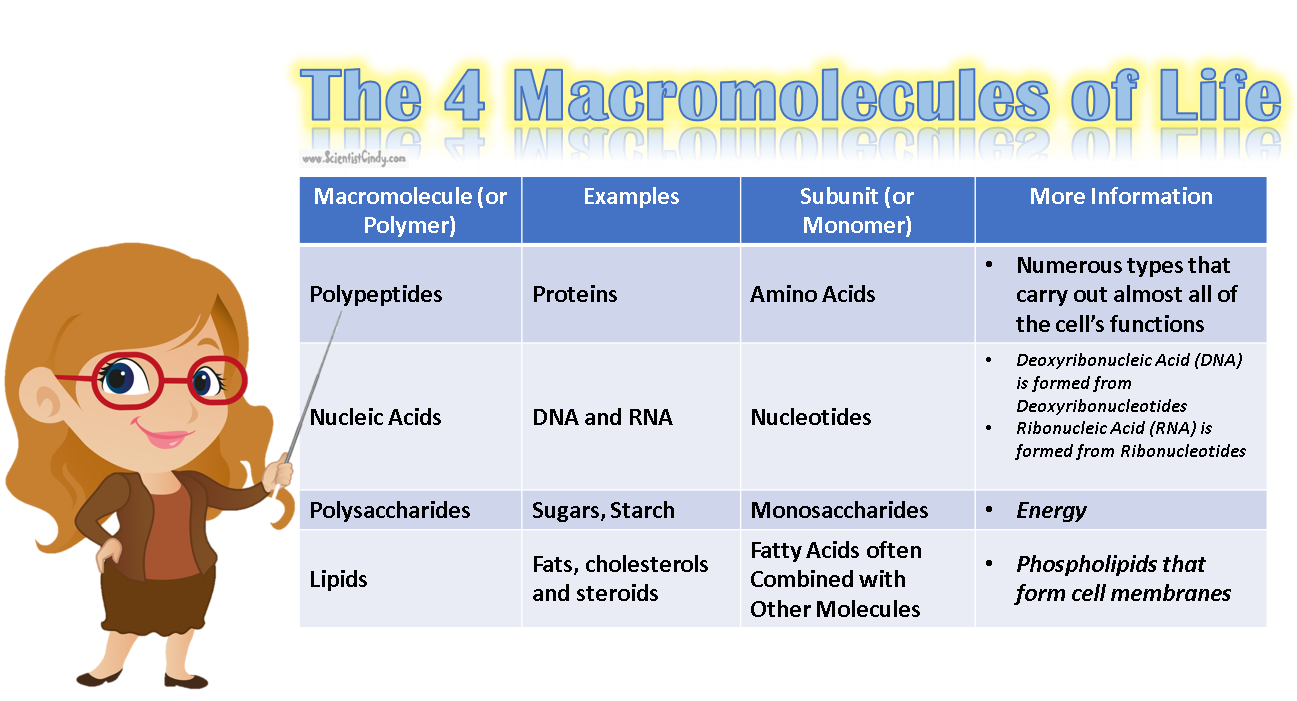
•Genes are part of genetic elements: large molecules and/or chromosomes. •Nucleic acids: contain genetic information via nucleotides (monomers of nucleic acids) •DNA (genetic blueprint) and RNA (transcription product) are polynucleotides. •Informational macromolecules include nucleic acids and proteins. |
DNA STRUCTURE
|
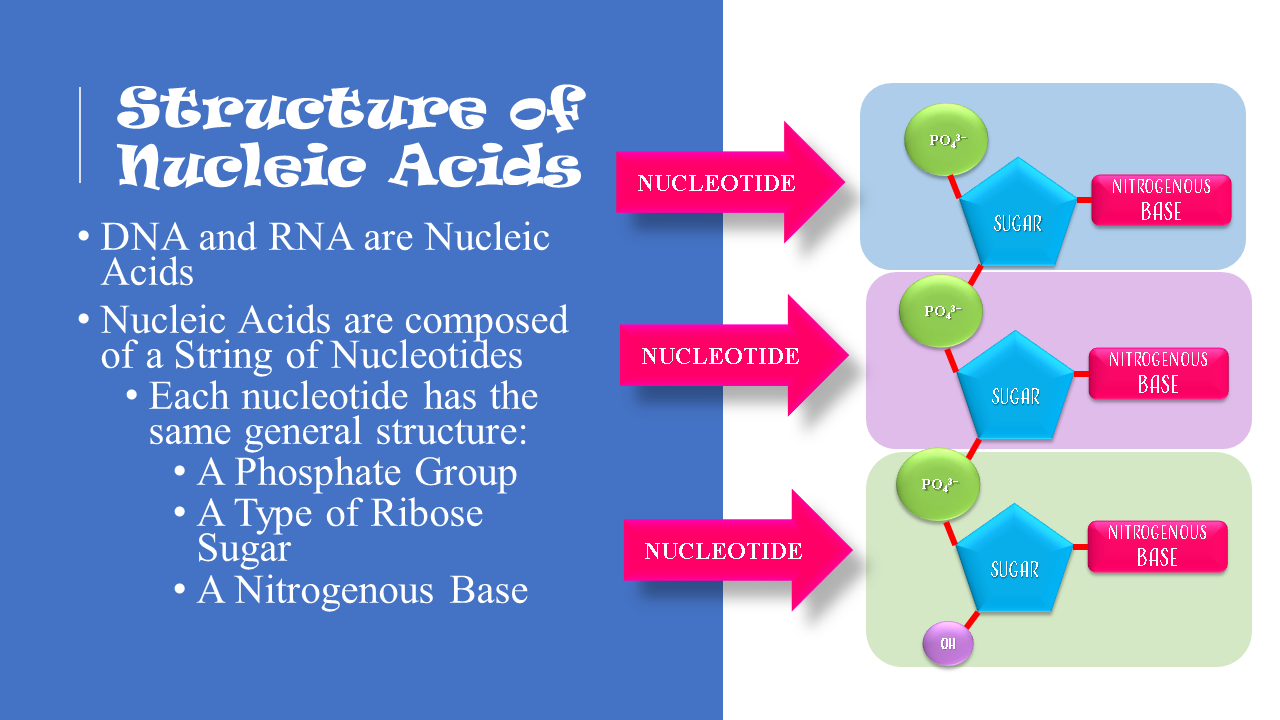
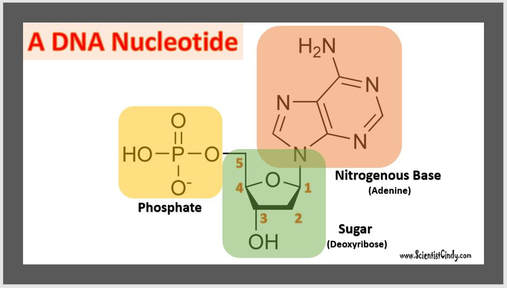
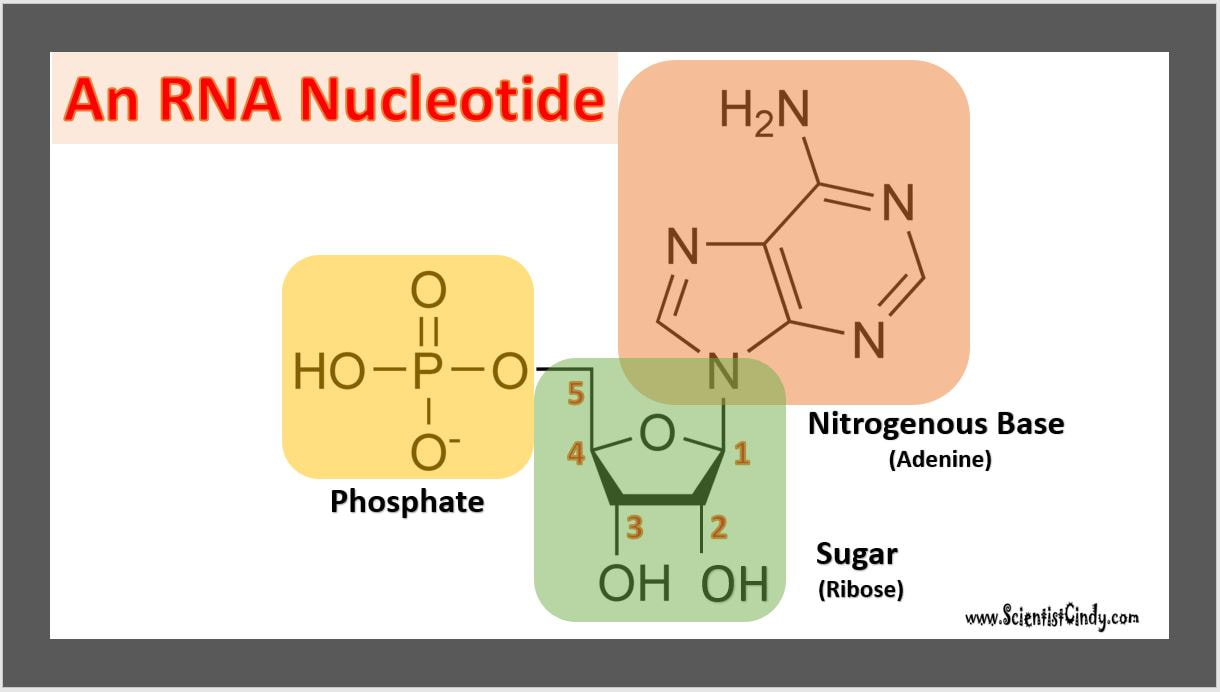
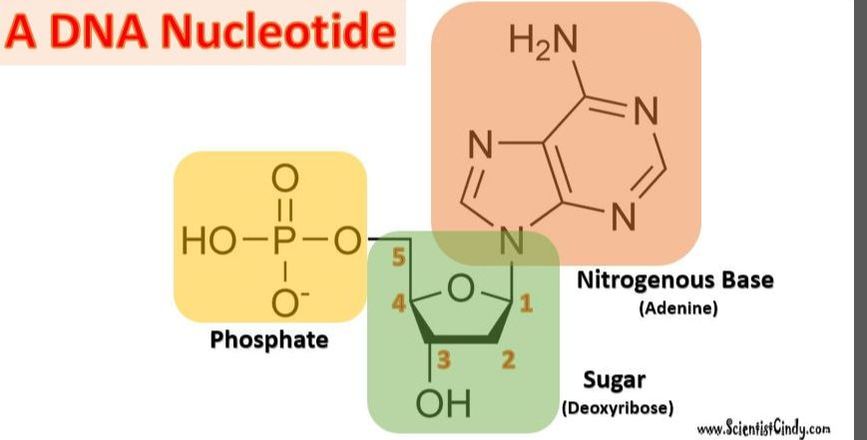
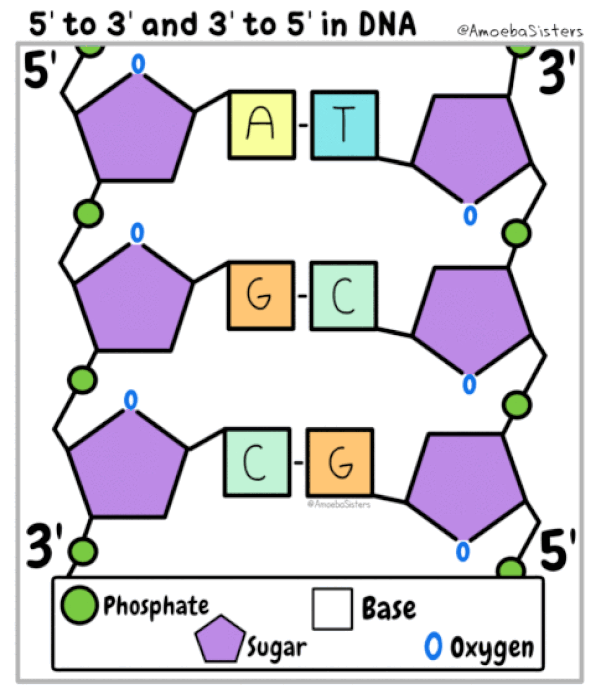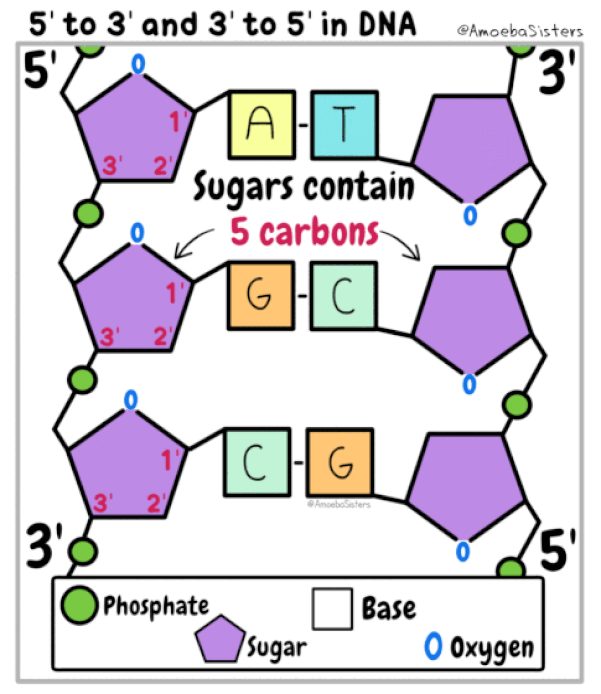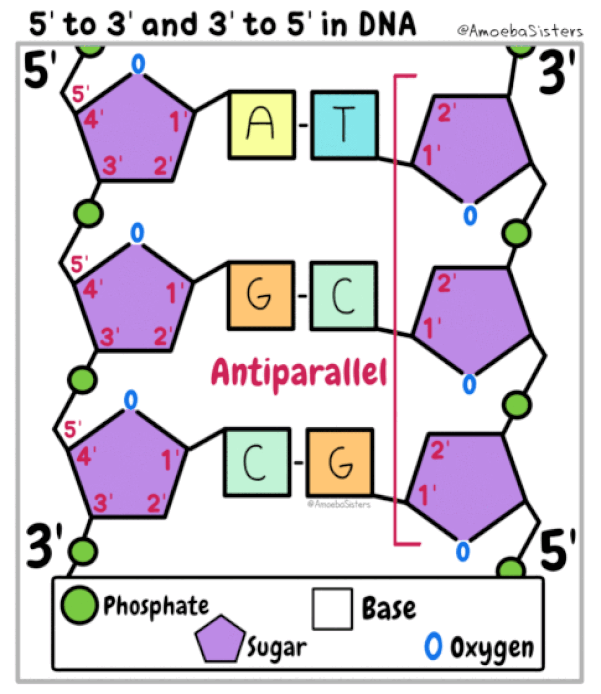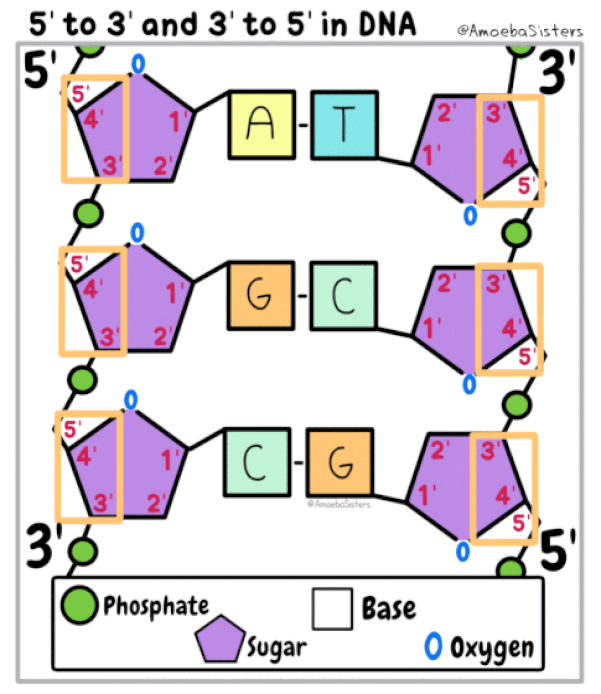
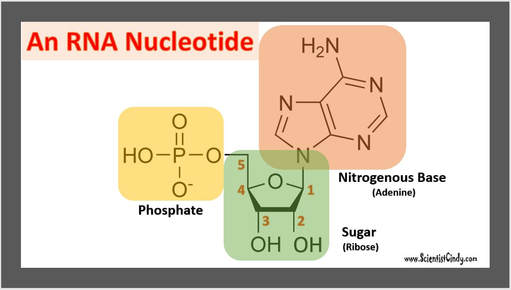
Nucleotides are the building blocks for DNA and RNA in living organisms. An individual nucleic acid is composed of the following:
|
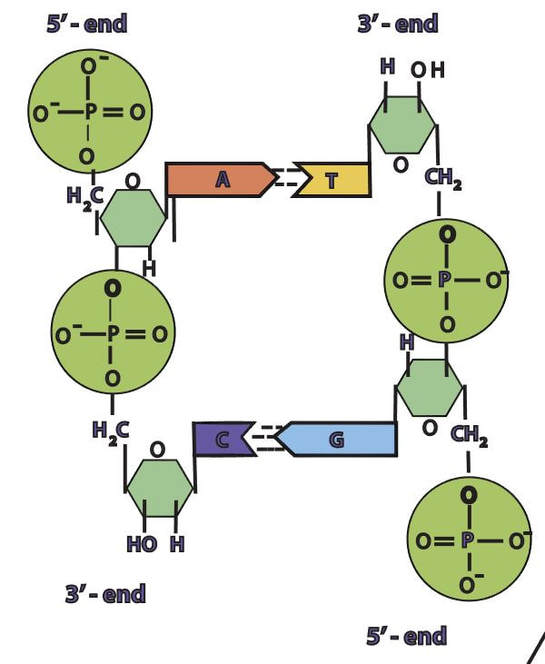
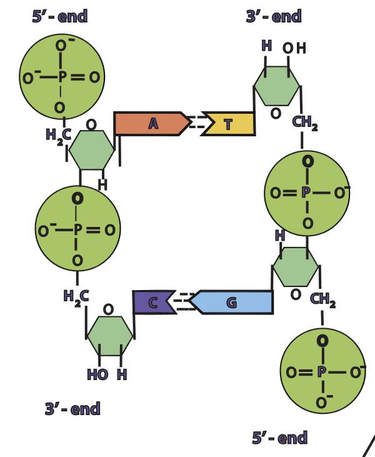
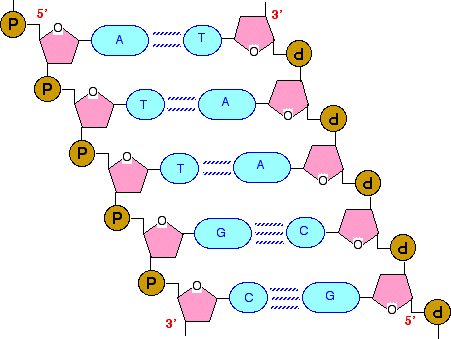
In a molecule of DNA or RNA, individual nucleotides are linked together to make a sugar-phosphate backbone. The backbone of DNA is made up of alternating deoxyribose sugar molecules and phosphate molecules bound together by phosphodiesterase bonds
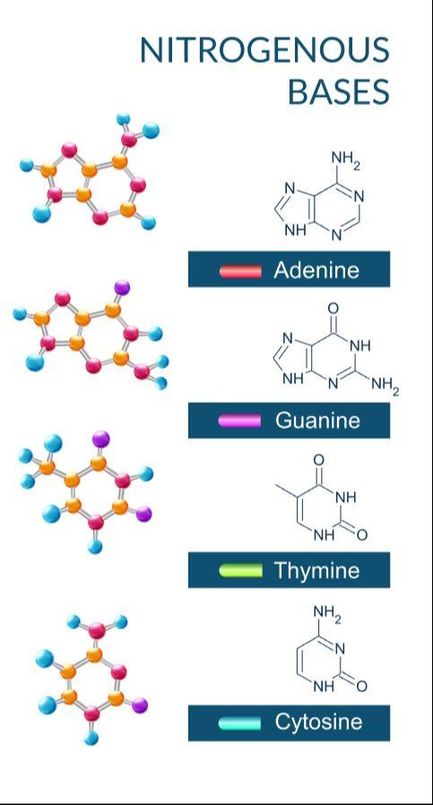
There are only 4 nitrogenous bases found in DNA
|
|
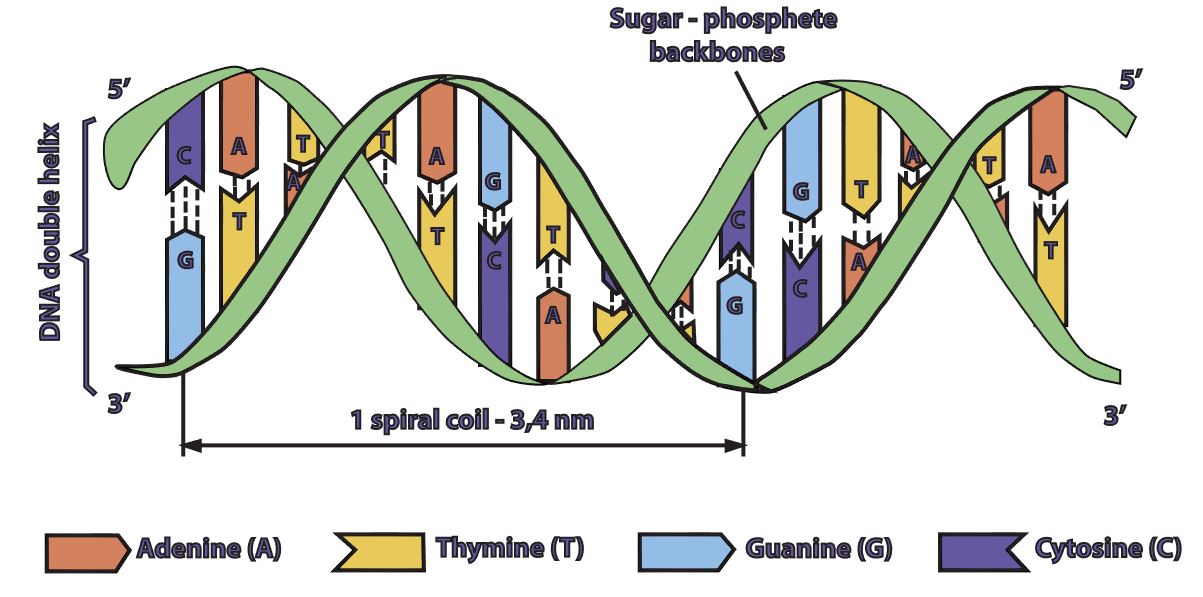
DNA is double stranded. Each strand of DNA is connected to the other DNA strand though hydrogen bonding. This hydrogen bonding occurs between the nitrogenous bases on opposite DNA strands.
|
|
The DNA strands are oriented antiparallel to one another. The ends of the DNA molecule are different. One end of the DNA molecule is designated 3' (stated as 3 prime) and the other as 5' (stated as 5 prime). They are names according to the carbons in the ribose sugar molecule of the DNA backbone. In a molecule of DNA, one strand will be oriented from 3' to 5', and the other from 5' to 3'.
|
All Living Organisms Use
DNA as Their Genetic Material.
What is Considered "Alive"?
Prokaryotes, like bacteria, and eukaryotes are considered to be living organisms. The current criteria for life includes that all lifeforms must consist of one or more cells. Viruses are not made up of cells, so they are not currently considered to be living organisms. Many scientists believe the criteria for life is outdated and should include viruses. Another criterion of life is that all lifeforms must have a means of reproduction. Viruses do have the ability to reproduce, but only with the assistance of a host cell. Viral genetics and reproduction strategies will be discussed later on this page.
Genetics in Eukaryotic Animal Cells vs. Bacteria
|
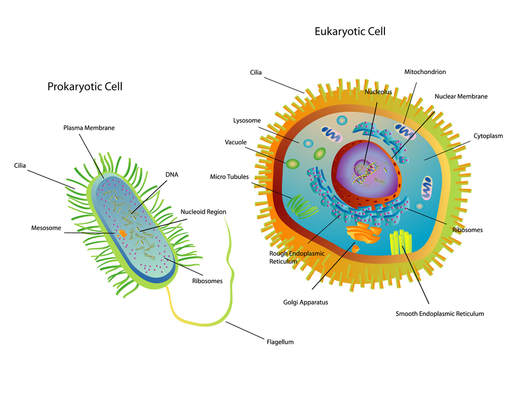
In eukaryotic cells, the genetic material (DNA) is in the nucleus of the cell. DNA holds the instructions for all of the proteins the cell may required. These instruction are encoded as the sequence of base pairs on the individual nucleic acids that make up the cell.
|
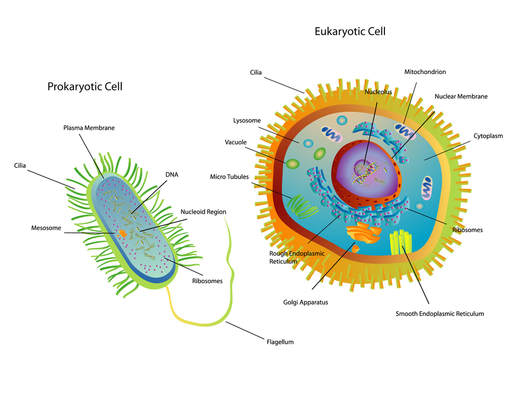
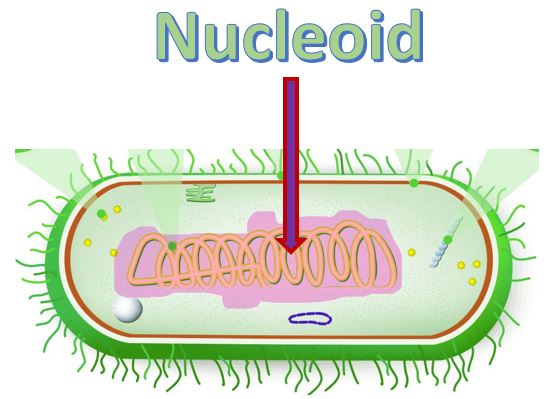
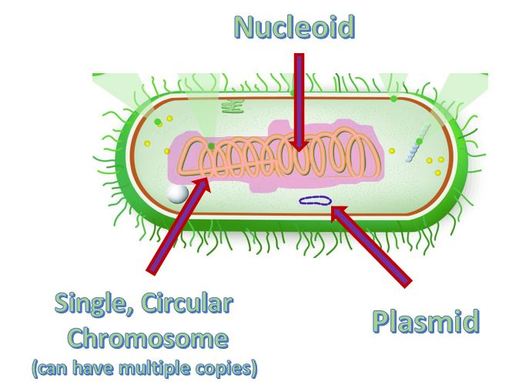
Bacteria cells are prokaryotic and are much smaller than the eukaryotic cell. They do not have any membrane-bound organelles. Also, bacteria cells do not have a nucleus, but they do have a region called the nucleoid, which is a region in which the DNA can be found.
|
Both bacteria and eukaryotes (animals, plants fungi and algae) are able to reproduce. We will narrow our focus by comparing animal cells specifically (as an example of eukaryotic cells) with bacteria cells.
In order for reproduction to occur, the DNA must be replicated. This process is simply called DNA Replication.
In order for reproduction to occur, the DNA must be replicated. This process is simply called DNA Replication.
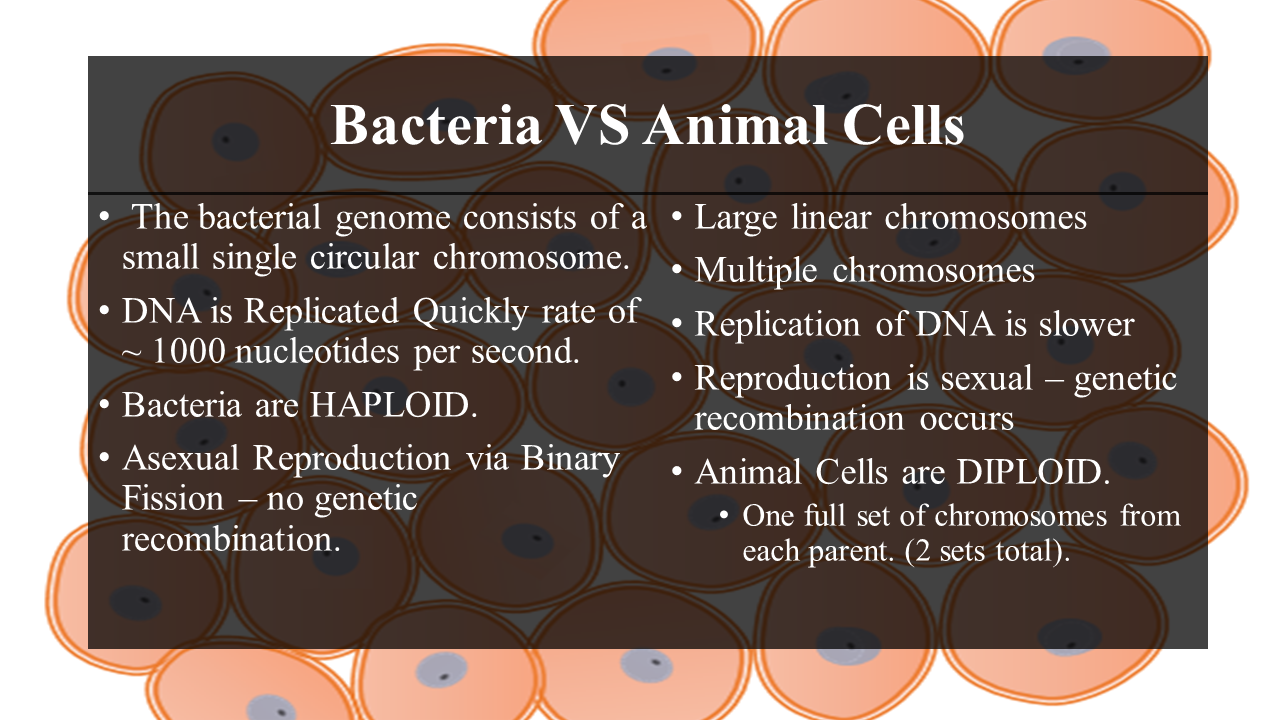
Prokaryotic cells have no nucleus, no organelles and a small amount of DNA in the form of a single, circular chromosome.
- Eukaryotic cells on the other hand, have a nucleus, multiple organelles and more DNA arranged in multiple, linear chromosomes.
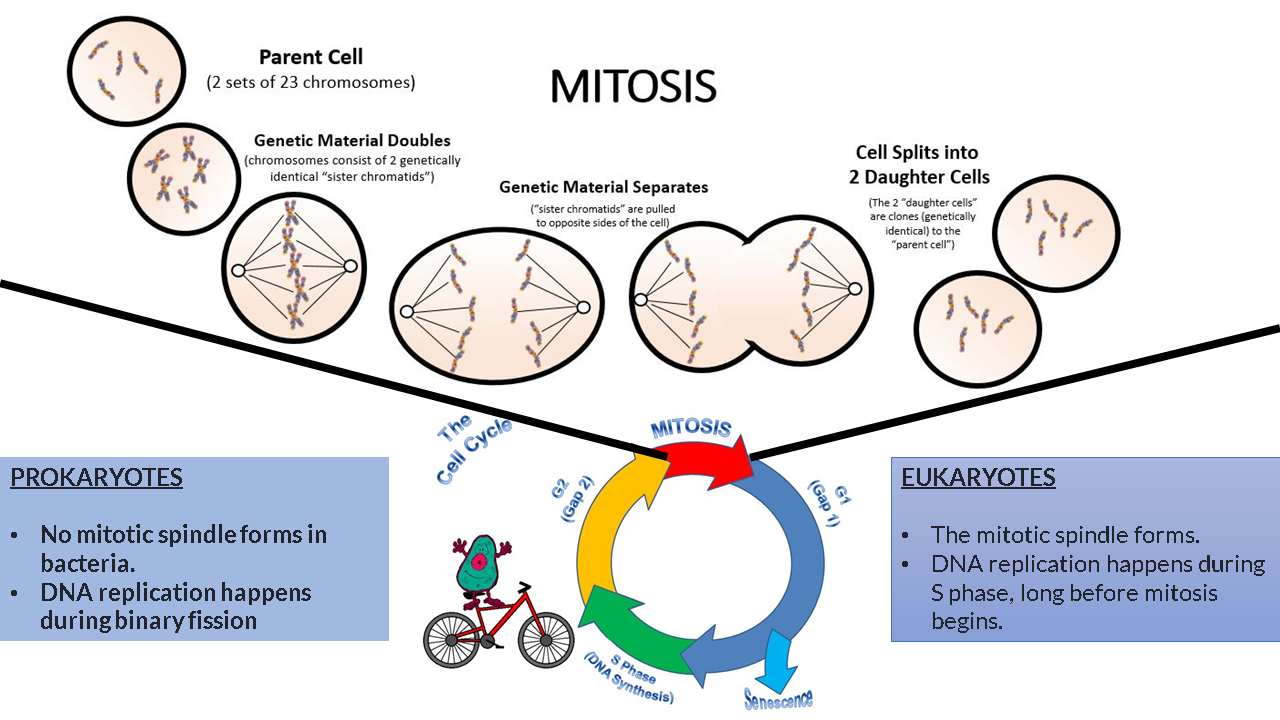
CELL DIVISION:
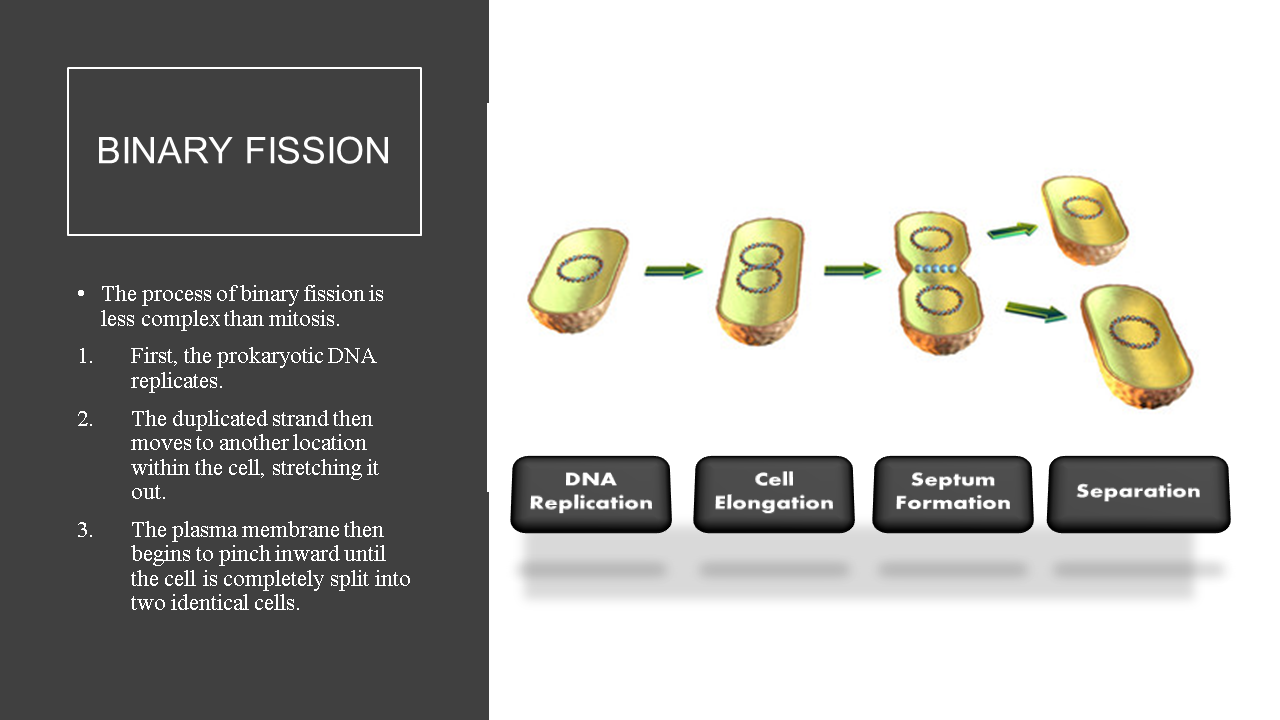
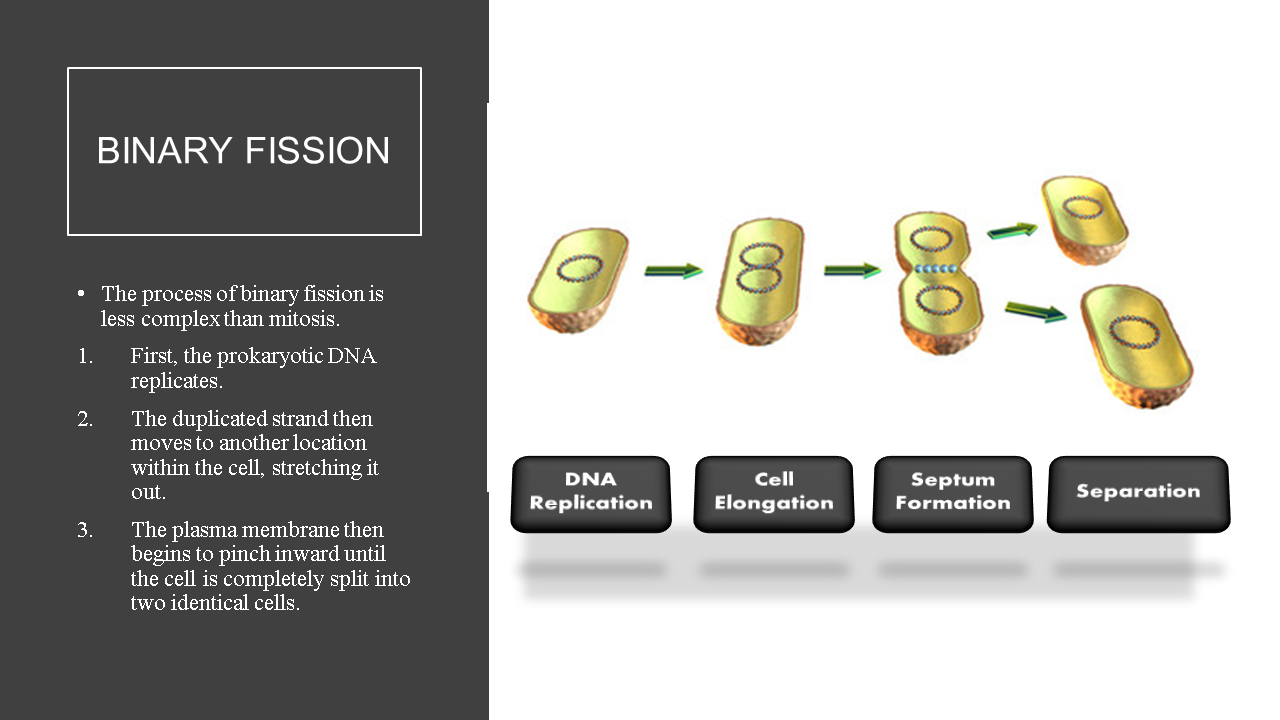
- Prokaryotic cells undergo cell division using binary fission; an asexual process that produces 2 genetically identical daughter cells from a single parent cell.
- prokaryotic cells are haploid; they have only one copy of their genome.
Eukaryotic cells undergo cell division using either the process of mitosis or meiosis.
- mitosis is an asexual process that produces 2 genetically identical daughter cells from a single parent cell. This process is performed by somatic cells. The parent cell and resulting daughter cells are diploid.
Parameter |
DNA REPLICATION IN BACTERIA |
DNA REPLICATION IN EUKARYOTES |
SIZE OF GENOME |
Tiny (~50 times smaller than eukaryotic genomes) |
Huge (~50 times larger than bacteria genomes) |
CONFIGURATION |
One Single Circular Chromosome |
Multiple Linear Chromosomes |
LOCATION |
Occurs in the Cytoplasm |
Occurs in the Nucleus |
TIMING |
Occurs During the First Step of Binary Fission |
Occurs During the S-Phase (Synthesis Phase) of the Cell Cycle |
ORIGIN OF REPLICATION |
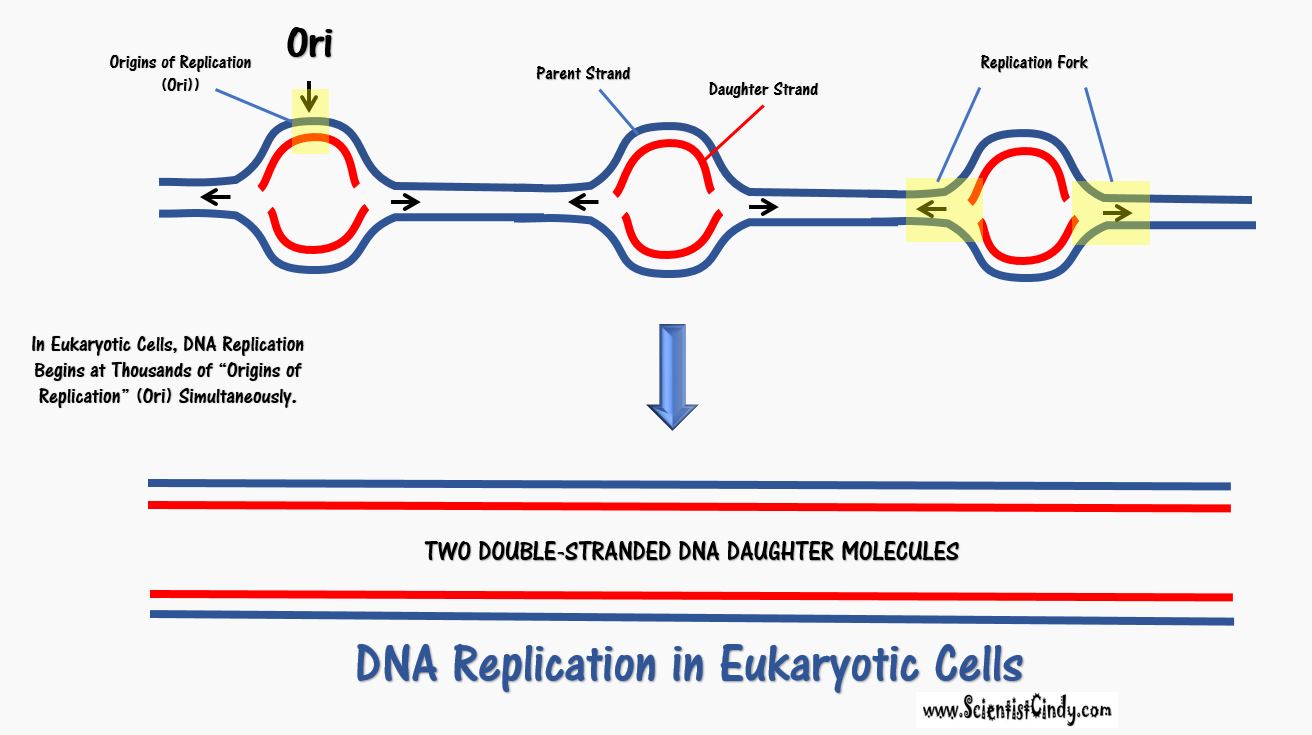
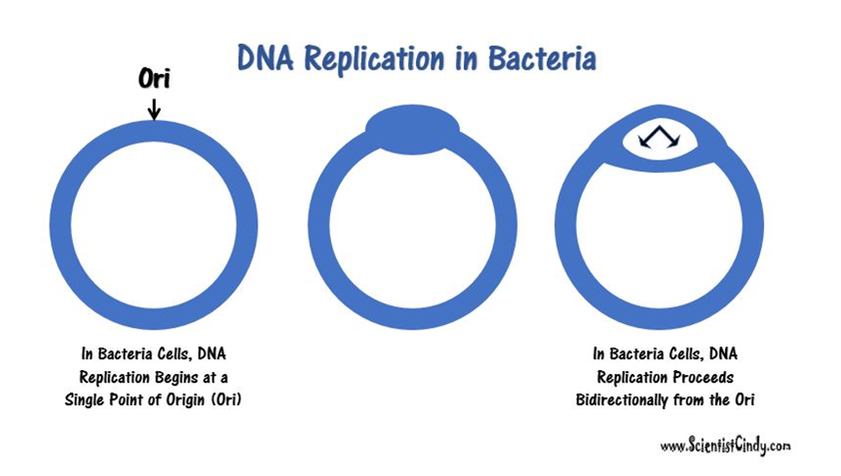
Begins at a Single Point of Origin. This is known as the "Origin of Replication" (Ori). |
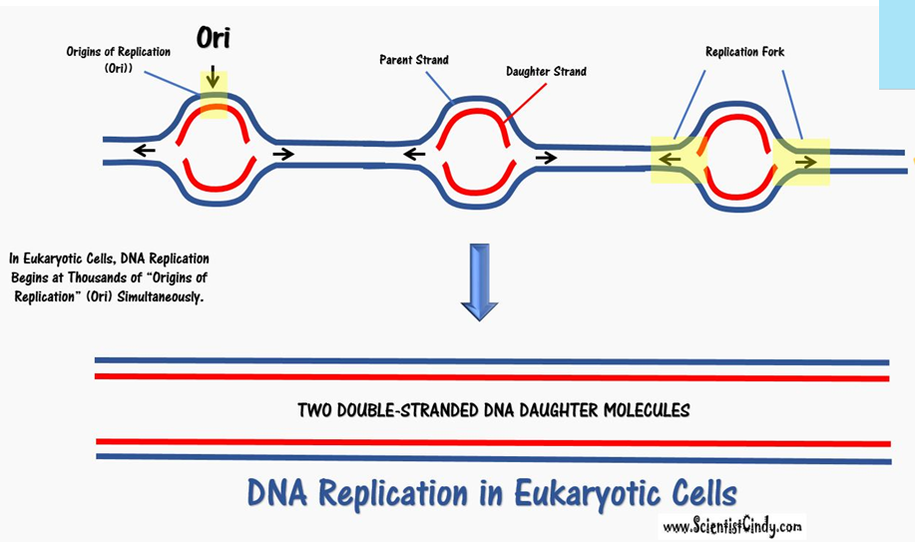
Begins at Thousands of Points of Origin. These are known as the "Origins of Replication" (Ori). |
DNA POLYMERASE |
DNA Polymerase III |
Many DNA Polymerases |
SPEED |
Faster (1000's of Nucleotides per Second) |
Slower (100's of Nucleotides per Second) |
TELOMERASE |
NO |
YES - telomeres are present |
Leading and Lagging Strand (Okazaki Fragments) |
YES - longer fragments |
YES - shorter fragments |
Value |
DNA Replication in Prokaryotic Cells
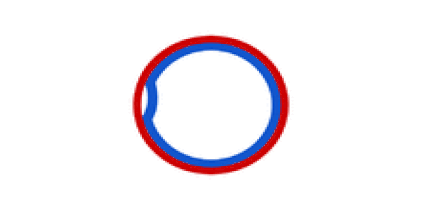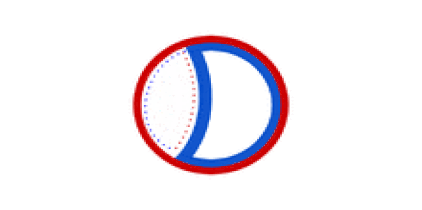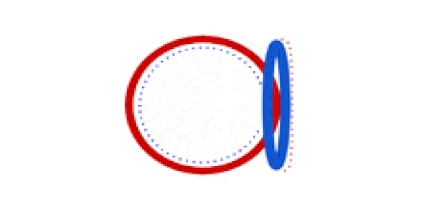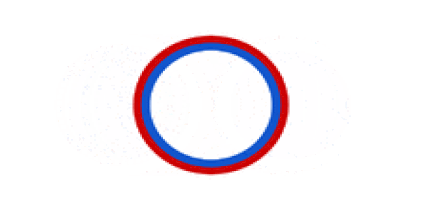
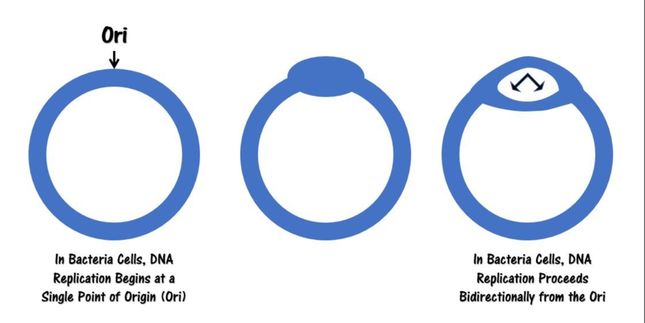
Circular bacterial chromosomes are replicated bidirectionally, starting at one point of origin and replicating in two directions away from the origin. This results in semi-conservative replication, in which each new identical DNA molecule contains one template strand from the original molecule, shown as the solid lines, and one new strand, shown as the dotted lines.
DNA Replication in Bacteria
(for Binary Fission)
|
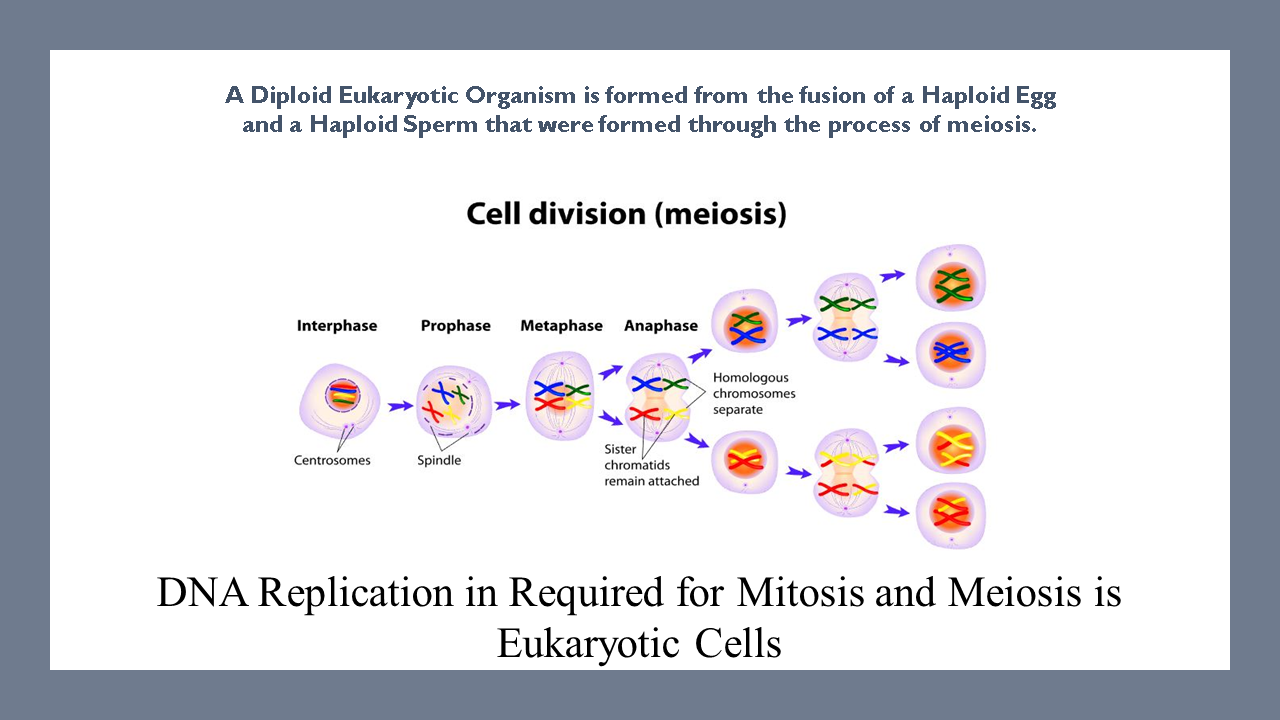
Eukaryotic cells carry two copies of each chromosome; one copy from each parent. Because eukaryotic cells have 2 sets of chromosomes, they are considered diploid.
Diploid offspring are formed from the fusion of a haploid egg (carrying one complete set of genes from the mother) combining with a haploid sperm (carrying one complete set of genes from the father) during fertilization. This process creates genetically unique offspring.
|
|
|
Bacterial cells reproduce using binary fission, which is an asexual process.
Since there is only one copy of the chromosome, bacterial cells are considered haploid. Bacteria cells are haploid which means they contain a single set of genes that they inherit from a single parent cell through the asexual process of reproduction known as binary fission.
The bacterial genome consists of a small single circular chromosome. The entire bacterial genome can be replicated in under an hour, at a rate of ~ 1000 nucleotides per second.
|
|
The DNA in bacteria exist in a region called the nucleoid. The nucleoid is simply a "cytoplasmic location" and not a membrane-bound organelle, like our nucleus. However, the nucleoid does succeed in packaging the DNA into a small space within the cytoplasm. Proteins assist in anchoring the genetic material to the nucleoid. Bacteria are small, and the nucleoid takes up almost 1/3 of the space in the cell!.
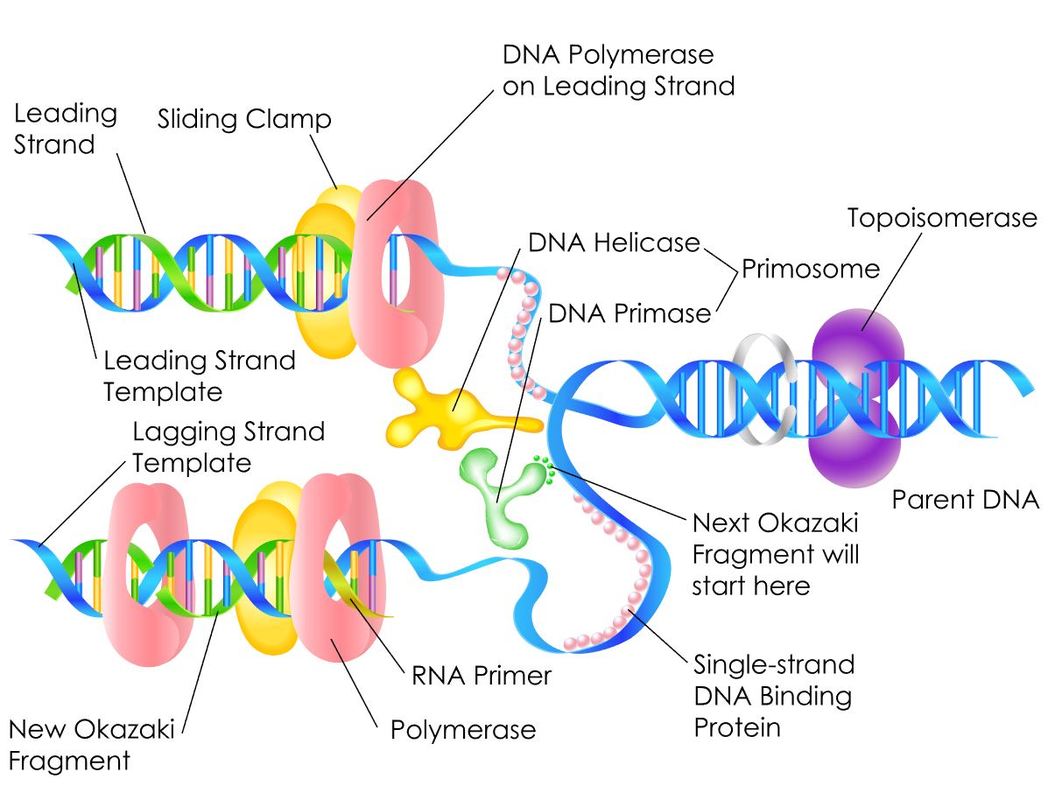
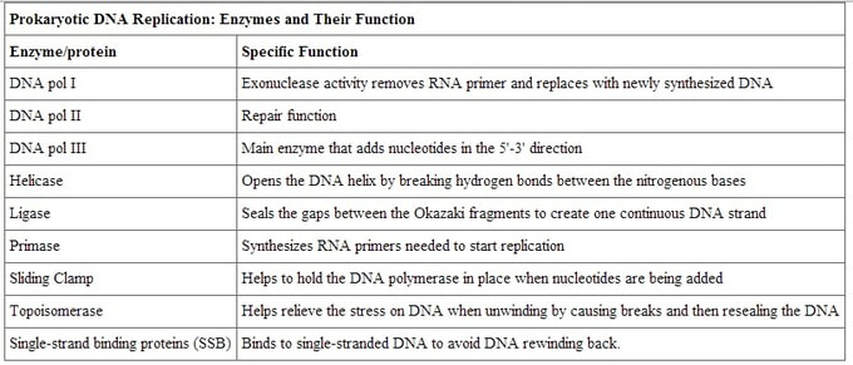
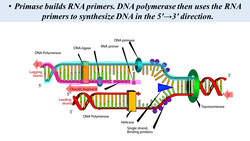
Helicase separates the DNA to form a replication fork at the origin of replication, Replication forks extend bi-directionally as replication continues. Okazaki fragments are formed on the lagging strand, while the leading strand is replicated continuously. DNA ligase seals the gaps between the Okazaki fragments. Primase synthesizes an RNA primer with a free 3′-OH, which DNA polymerase III uses to synthesize the daughter strands.
|
|
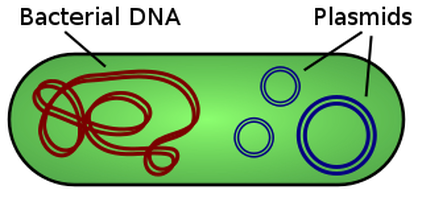
Some species of bacteria have an additional small extra-chromosomal circular DNA loop in their cytoplasm, called plasmids. Interestingly, these plasmids do not carry any of the genes essential for normal life processes, but they do alter the sensitivities or abilities of the bacteria. These non-essential genes are classified as accessory genes. For example, some plasmids carry genes that allow for antibiotic resistance, habitat tolerance or bacterial conjugation which allows the exchange of genetic material between bacterial cells.
DNA Synthesis / Genetic Replication
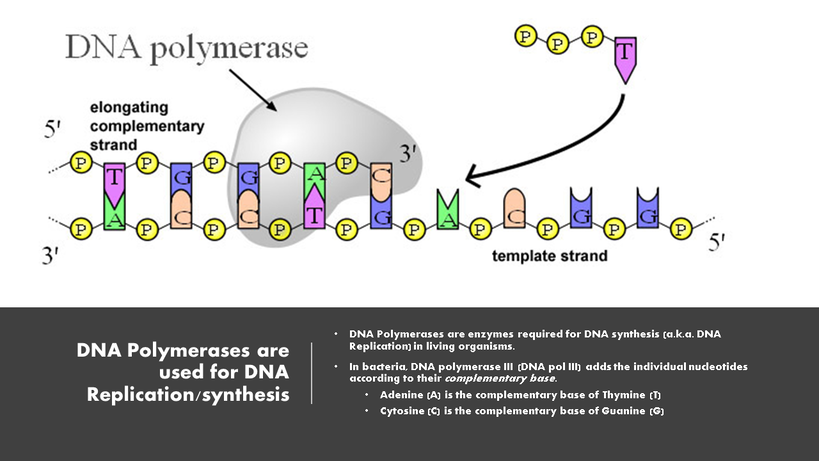
DNA pol I and DNA pol II are primarily required for repair, while DNA pol III is the enzyme required for DNA replication in bacteria.
DNA Pol III
|
DNA Pol III is the enzyme required for DNA synthesis in bacteria. DNA pol III adds the individual nucleotides according to their complementary base.
Remember that the rules for base-pairing of the nitrogenous bases of the nucleotides that make up DNA are as follows:
|
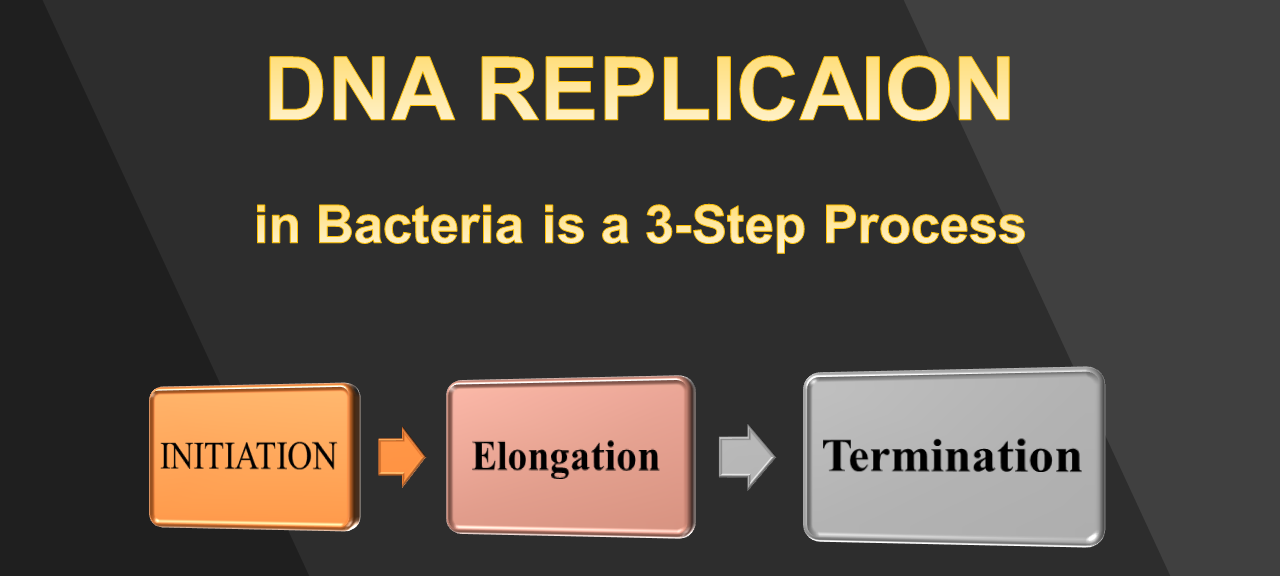
Step One - INITIATION (of DNA Replication)
Steps of INITIATION:
- DNA Replication in Prokaryotes:
- A replication fork is formed when helicase separates the DNA strands at the origin of replication.
- Topoisomerase I and II break and reform DNA’s phosphate backbone ahead of the replication fork, thereby relieving the pressure that results from this supercoiling. The DNA tends to become more highly coiled ahead of the replication fork. SEE VIDEO!
- Single-strand binding proteins bind to the single-stranded DNA to prevent the helix from re-forming.
- Primase synthesizes an RNA primer.
• There are specific nucleotide sequences called origins of replication (Ori) located on the bacterial chromosome.The Ori is the point at which DNA replication begins. Prokaryotic organisms will have a single origin of replication. Eukaryotic DNA can be about 50 times larger than the prokaryotic genome. Since so much DNA must be copied in the eukaryotic cell, we have thousands of origins of replication (Ori) all working at the same time.
DNA polymerase is able to add nucleotides only in the 5′ to 3′ direction (a new DNA strand can be extended only in this direction). It also requires a free 3′-OH group to which it can add nucleotides by forming a phosphodiester bond between the 3′-OH end and the 5′ phosphate of the next nucleotide.
RNA primase, synthesizes an RNA primer that is complementary to a short sequence of DNA. The RNA primer provides the free 3′-OH end needed to allow replication to occur. RNA pol III then begins adding nucleotides one by one that are complementary to the template (parent) DNA strand.
RNA primase, synthesizes an RNA primer that is complementary to a short sequence of DNA. The RNA primer provides the free 3′-OH end needed to allow replication to occur. RNA pol III then begins adding nucleotides one by one that are complementary to the template (parent) DNA strand.
A primer is a short single strand of RNA or DNA (generally about 18-22 bases) that serves as a starting point for DNA synthesis. It is required for DNA replication because the enzymes that catalyze this process, DNA polymerases, can only add new nucleotides to an existing strand of DNA. The polymerase starts replication at the 3′-end of the primer, and copies the opposite strand.
DNA gyrase, a.k.a. gyrase, or topoisomerase II relieves strain while double-stranded DNA is being unwound by helicase. The enzyme does this by cutting one of the double helices. This enzyme is especially important for prokaryotes, because the circular chromosome gets super-coiled during replication.
Step Two - ELONGATION (of DNA Replication)
ELONGATION - After initiation, the DNA is ready to be replicated. This occurs quickly in the bacterial cell (approximately 1000 nucleotides per second). The main idea of elongation is that the DNA Polymerase III will add on complimentary nucleotides (according to the rules of base-pairing).
DNA Polymerase III can only synthesize DNA in the 5’ to 3’ direction. This fact creates a difficult situation. You see, DNA is double stranded with each of those strands being reversed or antiparallel. In other words, one strand is oriented in the 5’ to 3’ direction and the other is oriented in the 3’ to 5’ direction. The consequence of this antiparallel configuration, coupled with the factor that DNA Pol III can only synthesize DNA in the 5' to 3' direction, is that one of the DNA strands will be replicated continuously, but the other will not!
DNA Polymerase III can only synthesize DNA in the 5’ to 3’ direction. This fact creates a difficult situation. You see, DNA is double stranded with each of those strands being reversed or antiparallel. In other words, one strand is oriented in the 5’ to 3’ direction and the other is oriented in the 3’ to 5’ direction. The consequence of this antiparallel configuration, coupled with the factor that DNA Pol III can only synthesize DNA in the 5' to 3' direction, is that one of the DNA strands will be replicated continuously, but the other will not!
Leading Strand - The parent strand of DNA which runs in the 3' to 5' direction toward the replication fork. It has the ability to be replicated continuously by DNA polymerase.
Lagging Strand - This is the parent strand of DNA which runs in the 5' to 3' direction toward the fork, and it's replicated discontinuously.
Lagging Strand - This is the parent strand of DNA which runs in the 5' to 3' direction toward the fork, and it's replicated discontinuously.
- The lagging strand of DNA is that strand of the DNA double helix that is orientated in a 5′ to 3′ manner. Therefore, its complement must be synthesized in a 3′→5′ manner. Because DNA polymerase III cannot synthesize in the 5′→3′ direction, the lagging strand is synthesized in short segments known as Okazaki fragments. Along the lagging strand's template, primase builds RNA primers in short bursts. DNA polymerases are then able to use the free 3′-OH groups on the RNA primers to synthesize DNA in the 5′→3′ direction.
Steps of Elongation:
- DNA polymerase III uses the RNA primer to synthesize the daughter DNA strand.
- On the leading strand, DNA is synthesized continuously, whereas on the lagging strand, DNA is synthesized discontinuously as Okazaki fragments.
- On the Leading Strand, DNA Polymerase III follows helicase and adds on nucleotides according to complementary bases in the 3' to 5' direction. This is considered continuous replication.
- On the Lagging Strand, DNA Polymerase III will replicate short portions of DNA at a time, called Okazaki Fragments. This is considered discontinuous replication.
- On the leading strand, DNA is synthesized continuously, whereas on the lagging strand, DNA is synthesized discontinuously as Okazaki fragments.
- DNA polymerase I replaces the RNA primer with DNA.
- The RNA fragments are then removed by DNA polymerase I for prokaryotes or DNA polymerase δ for eukaryotes and new deoxyribonucleotides are added to fill the gaps where the RNA was.
- DNA ligase seals the gaps between the Okazaki fragments, joining the fragments into a single DNA molecule.
- DNA Ligase is the enzyme that functions to connect the Okazaki Fragments together for the copy of the lagging strand. The word "ligate" in Latin means "to tie together."
- DNA ligase then joins the deoxyribonucleotides together, completing the synthesis of the lagging strand.
- DNA Ligase is the enzyme that functions to connect the Okazaki Fragments together for the copy of the lagging strand. The word "ligate" in Latin means "to tie together."
Step Three - TERMINATION (of DNA Replication)
- TERMINATION - After the full chromosome has been replicated, termination of DNA replication must occur. Since the bacterial genome consists of a single circular chromosome, at the end of the elongation step of DNA replication, you end up with 2 interlocking circles of DNA. The scientific terms, you would say that the parent DNA and the daughter DNA molecules are concatenated., These concatenated DNA chromosomes must be separated.
Steps of Termination in prokaryotic cells
- Bacterial Topoisomerase IV induces a double-strand break into the DNA molecules, allowing them to separate from each other (becoming un-concatenated).
- Bacterial Topoisomerase IV then reseals each DNA molecule back together again creating 2 separate, un-linked chromosomes.
Telomeres
- telomere: the repetitive nucleotide sequences at each end of a eukaryotic chromosome, which protect the chromosome from degradation following many rounds of replication
- telomerase: an enzyme in eukaryotic cells that adds a specific sequence of DNA to the telomeres of chromosomes after they divide which rebuilds the telomeres after cell division in some eukaryotic cells.
The chromosomes of eukaryotes are linear, which causes an issue with the termination step of DNA replication.
The trouble is that the ends of these linear chromosomes cannot be fully replicated. This means that every time a eukaryotic cell undergoes cell division, the chromosome becomes increasingly shorted with each round of DNA replication
The trouble is that the ends of these linear chromosomes cannot be fully replicated. This means that every time a eukaryotic cell undergoes cell division, the chromosome becomes increasingly shorted with each round of DNA replication
A few difference to note: The basic steps are pretty much the same in bacteria vs. eukaryotic cells, but the main difference is that eukaryotic chromosomes are linear. The ends of linear chromosomes have specialized non-coding repeated sequences of DNA called telomeres. The function of the telomeres is to protect the valuable coding sequences of the DNA strand from being lost. You see, in eukaryotic cells, every time a cell divides, the DNA is replicated, Every time the DNA is replicated, there is a small section of DNA at the end (the telomeres) that are not able to be replicated,
Eukaryotic chromosomes have adapted a mechanism to deal with the progressive loss of genetic information at the end of linear chromosomes, by creating long, repetitive sequences called telomeres, which exist at the chromosome ends. These telomeres may have hundreds or thousands of repeats that do not code for proteins.
Telomerase
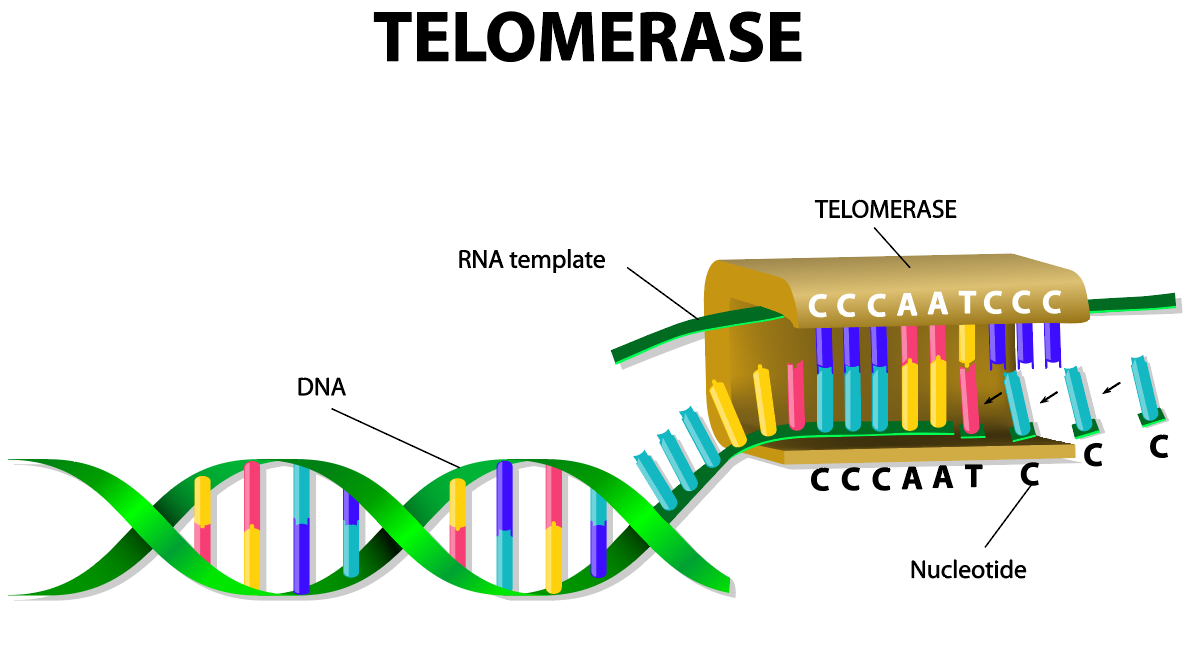
Some cells have the ability to reverse telomere shortening using telomerase, an enzyme that extends the telomeres of chromosomes. The enzyme telomerase has a built-in RNA template so it can add on more of the repeated segments of DNA to the ends of the telomeres. In humans, telomerase is typically active in eggs and sperm cells (gametes) and adult stem cells, but it is not active in other cells. It is currently believed that telomere shortening causes signs of aging of these cells.
Steps in DNA Replication
- DNA replication begins at a specific spot on the DNA molecule called the origin of replication.
- At the origin, enzymes unwind the double helix making its components accessible for replication. Each strand of the helix then separates from the other, exposing the now unpaired bases to serve as templates for new strands. A small segment of RNA is added as a primer, then new nucleotide bases that complement the unpaired bases can be assembled to form two daughter strands next to each parent strand. This assembly is accomplished with enzymes called DNA polymerases. When the process is complete, two DNA molecules have been formed identical to each other and to the parent molecule.
The steps for DNA replication are generally the same for all prokaryotic and eukaryotic organisms.
- Unwinding the DNA is accomplished by an enzyme named DNA helicase.
- DNA replication is initiated at the Ori and form a replication bubble with replication forks traveling in both directions from the Ori
- Both eukaryotic and prokaryotic organisms use DNA polymerases for DNA replication.
- The types of RNA polymerases used are structurally different, but the function is very similar.
- DNA polymerase adds nucleotides to the 3' end of a strand of DNA in the 5' to 3' direction
- DNA polymerases require the presence of an RNA primer which is paired with a template strand.
- Semi-conservative replication.
- Both types of organisms also begin new DNA strands with a small primer of RNA.
- In this pattern, the individual strands of DNA are manufactured in different directions, producing a leading and a lagging strand.
- Lagging strands are created by the production of small DNA fragments called Okazaki fragments that are eventually joined together.
- The average eukaryotic cell has a lot more DNA than a prokaryotic cell.
- In prokaryotic cells, there is only one point of origin
- Eukaryotic cells have multiple points of origin
- Eukaryotic cells undergo DNA replication in the nucleus of the cell.
- Prokaryotic cells undergo DNA replication in the cytoplasm (or nucleoid)
- Eukaryotic chromosomes experience telomere shortening due to the linear nature of their chromosomes. Prokaryotic cells do not contain telomeres.
- Prokaryotic cells end up with 2 concatenated circles of DNA near the end of DNA replication
GENETIC RECOMBINATION IN BACTERIA
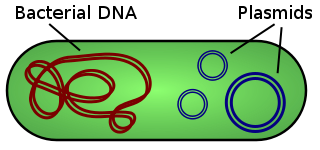
Bacteria contain a single circular chromosome which can be found in a general region called the nucleoid. Bacteria cells do not have a nucleus and do not contain any membrane-bound organelles.
Bacteria only reproduce asexually through a process called binary fission which produces genetic clones. There is no genetic diversity (or genetic recombination) introduced in bacterial cell division!
In addition to the single circular chromosome, some bacteria may contain additional small pieces of DNA called plasmids. Plasmids are small circular pieces of DNA that can contain between 1 and 10 genes (or so). Plasmids are going to be very important in terms of genetic recombination that takes place in bacteria. Bacteria have the ability to transfer these plasmids from one bacteria to another bacteria in order to create genetic variation.
Bacteria only reproduce asexually through a process called binary fission which produces genetic clones. There is no genetic diversity (or genetic recombination) introduced in bacterial cell division!
In addition to the single circular chromosome, some bacteria may contain additional small pieces of DNA called plasmids. Plasmids are small circular pieces of DNA that can contain between 1 and 10 genes (or so). Plasmids are going to be very important in terms of genetic recombination that takes place in bacteria. Bacteria have the ability to transfer these plasmids from one bacteria to another bacteria in order to create genetic variation.
Bacteria reproduce very quickly using the asexual process of binary fission. When we refer to a reproductive process as "asexual" this means that there is no exchange of genetic material. The offspring will be genetic clones of the parent cell and no genetic variation will occur through this process. Genetic variation increases the survival of the species through the process of natural selection. Natural selection is the method by which evolution takes place.
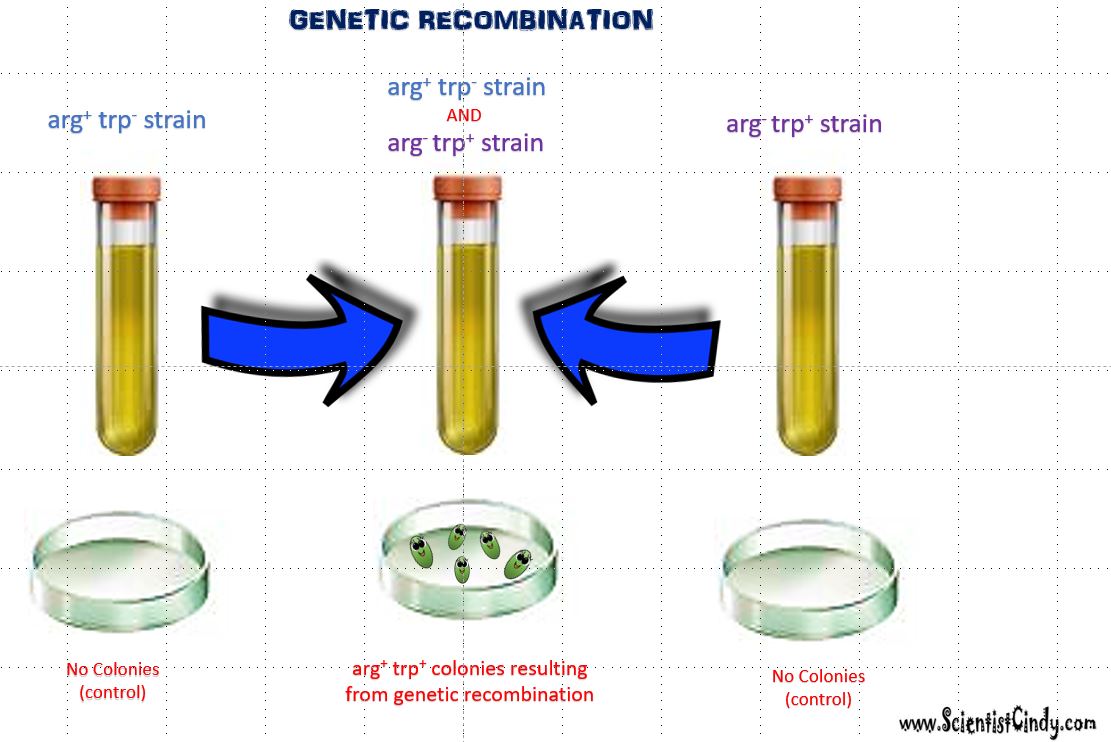
We see evidence of genetic recombination, when we are able to observe bacterial growth under conditions in which growth should not have occurred. Let's break down an example of this idea.
Here is the necessary background information that is important to know in order to understand the example below. When bacteria grows on a Petri dish, they form colonies. A single colony consists of thousands of bacteria. The growth media used in this experiment lacks both arginine (arg) and tryptophan (trp). Both arginine and tryptophan are needed for bacterial growth. If a bacterial cell has the ability to make its own arginine AND tryptophan, then it will be able to grow (as colonies) on this growth media. However, a bacteria cell that lacks the ability to create either arginine (arg) or tryptophan (trp) will not be able to grow on this growth media.
Here is the necessary background information that is important to know in order to understand the example below. When bacteria grows on a Petri dish, they form colonies. A single colony consists of thousands of bacteria. The growth media used in this experiment lacks both arginine (arg) and tryptophan (trp). Both arginine and tryptophan are needed for bacterial growth. If a bacterial cell has the ability to make its own arginine AND tryptophan, then it will be able to grow (as colonies) on this growth media. However, a bacteria cell that lacks the ability to create either arginine (arg) or tryptophan (trp) will not be able to grow on this growth media.
In the illustration, the first bacterial strain (left) carries the gene to create arginine (arg+), but it does NOT have the gene that creates tryptophan (trp-). Since the bacteria lacks the gene needed to create tryptophan, it will not grow on this growth media (no colonies will form). The bacterial strain on the right has the gene for tryptophan ( trp+) but does NOT have the gene for arginine (arg-). Since this bacteria cannot make its own arginine, no colonies will form using this growth media. However, when the two strains (arg+ trp-) and (arg- trp+) are combined in the same test tube, colonies are able to form on the Petri dish. This could only occur if the bacteria had somehow possessed the genes needed to produce both arginine and tryptophan.
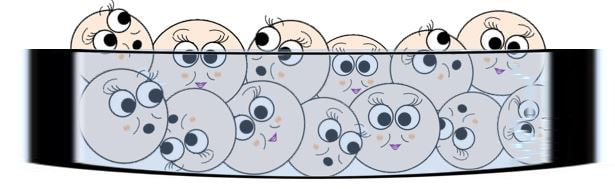 Illustration of arg+ trp+ colonies resulting from genetic recombintion
Illustration of arg+ trp+ colonies resulting from genetic recombintion
The two strains of bacteria, (arg+ trp-) and (arg- trp+), were able to form a new genetically unique strain of bacteria (arg+ trp+) that had the ability to make its own arginine (arg) and tryptophan (trp). Since this new strain of bacteria was able to synthesize its own arg and trp, it was able to grow on a Petri dish using medium lacking both the arginine and tryptophan that the bacteria requires for growth. The formation of this new strain involves the transference of genetic material between 2 individual bacteria cells from different strains.
There are 3 different methods by which genetic information can be exchanged in bacteria. Also known as horizontal gene transfer (HGT).
- TRANSFORMATION
- TRANSDUCTION
- CONJUGATION
2) TRANSFORMATION

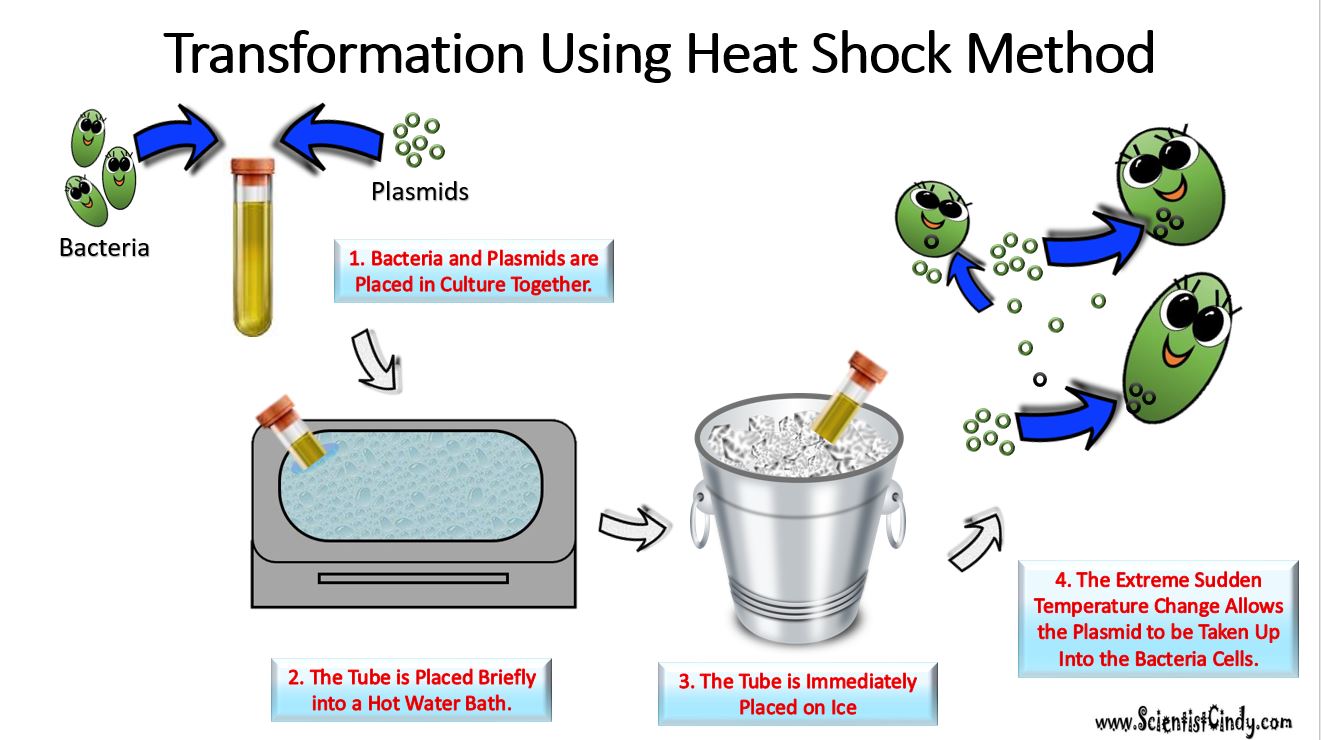
Transformation - Transformation is when a bacteria takes in plasmids from their environment. In order to take in a plasmid from the environment, microbiologists must "induce competency".
There are several methods used in laboratories to induce competency. One of the most popular and easiest ways to induce competency in bacteria, is the "heat shock" method. Heat shocking involves exposing the bacteria to a moderately high temperature (42 degrees Celsius is common) for a brief amount of time (usually no more than 45 seconds). Immediately following the heat exposure, the culture is then put on ice.
This extreme temperature change causes specialized surface proteins on the cell membrane to take up the extracellular plasmid. The bacteria are able to internalize the plasmids and they can express the genes on those plasmids.
There are several methods used in laboratories to induce competency. One of the most popular and easiest ways to induce competency in bacteria, is the "heat shock" method. Heat shocking involves exposing the bacteria to a moderately high temperature (42 degrees Celsius is common) for a brief amount of time (usually no more than 45 seconds). Immediately following the heat exposure, the culture is then put on ice.
This extreme temperature change causes specialized surface proteins on the cell membrane to take up the extracellular plasmid. The bacteria are able to internalize the plasmids and they can express the genes on those plasmids.
Natural transformation occurs in some bacterial species without the assistance of laboratory manipulation. There are currently 67 known bacterial species that are able to undergo this form of horizontal gene transfer. The ability depends on the expression of numerous bacterial genes whose products are responsible for this process. In general, transformation is a complex, energy-requiring developmental process.
In order for a bacterium to bind, take up and recombine exogenous DNA into its chromosome, it must become competent. To become "competent" the bacterium must enter a special physiological state. The DNA taken up from the environment will usually become integrated into its genome. This process is usually occurs among bacteria belonging to the same species, though exceptions have occurred.
In order for a bacterium to bind, take up and recombine exogenous DNA into its chromosome, it must become competent. To become "competent" the bacterium must enter a special physiological state. The DNA taken up from the environment will usually become integrated into its genome. This process is usually occurs among bacteria belonging to the same species, though exceptions have occurred.
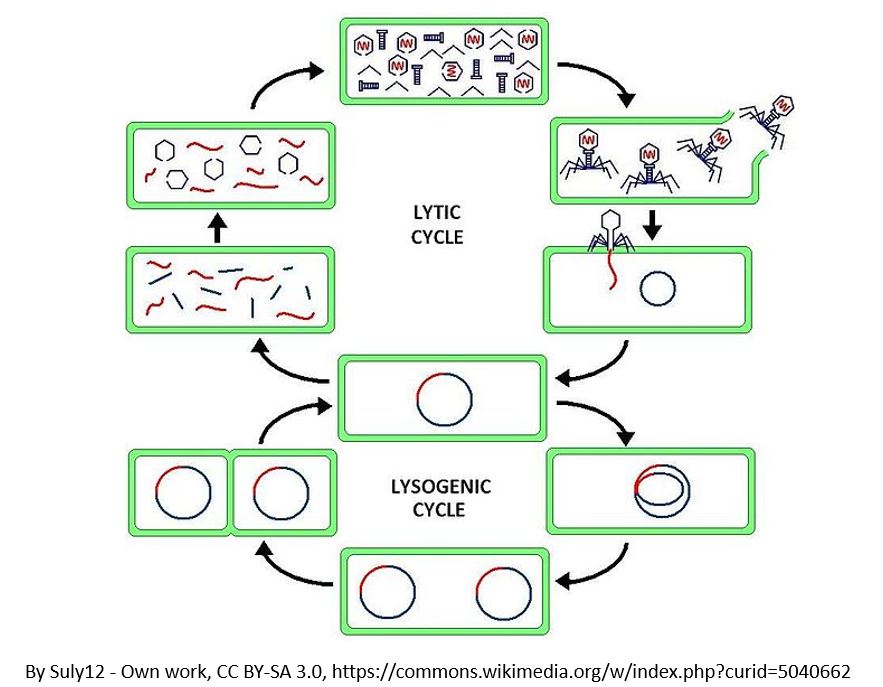
2) TRANSDUCTION
Phages can "accidentally" participate in horizontal gene transfer between bacteria cells. Viruses are able to
transfer genetic material when they infect a bacteria. This process is called transduction. DNA from the virus's previous host can be carried over to the newly infected cell. There are two
forms of transduction.
transduction, the phage is going to utilize the lysogenic cycle. In both of these processes, the phage "accidentally" transfers DNA from one bacteria to the next.
transfer genetic material when they infect a bacteria. This process is called transduction. DNA from the virus's previous host can be carried over to the newly infected cell. There are two
forms of transduction.
- Generalized Transduction
- Specialized Transduction
transduction, the phage is going to utilize the lysogenic cycle. In both of these processes, the phage "accidentally" transfers DNA from one bacteria to the next.
Steps of Generalized Transduction
- The phage is going to land on a bacteria cell (or bacterium).
- The phage inserts its genetic information
- The bacteria's machinery is used to form new phages
- The new phages leave the host cell.
- Occasionally, some phages may have some of the bacteria's DNA inside of it when the exit the host cell.
- In these cases, in which the phages are carrying bacteria DNA, these phages will brings that foreign bacterial DNA with it when it infects the next host cell.
- This time, when the phase goes to injects its DNA into the host cell, the bacterial DNA will be transferred to the "would-be" host cell instead!
Steps of Specialized Transduction
The process is the same as generalized except that the phage is going to utilize the lysogenic cycle.
The process is the same as generalized except that the phage is going to utilize the lysogenic cycle.
- The prophage is going to leave the bacteria cell,
- The prophage will contain a small piece of bacterial DNA
- When new viruses are assembled, bacterial DNA will be is some of them
- This bacterial DNA will get transferred to any new host cell the virus infects next.
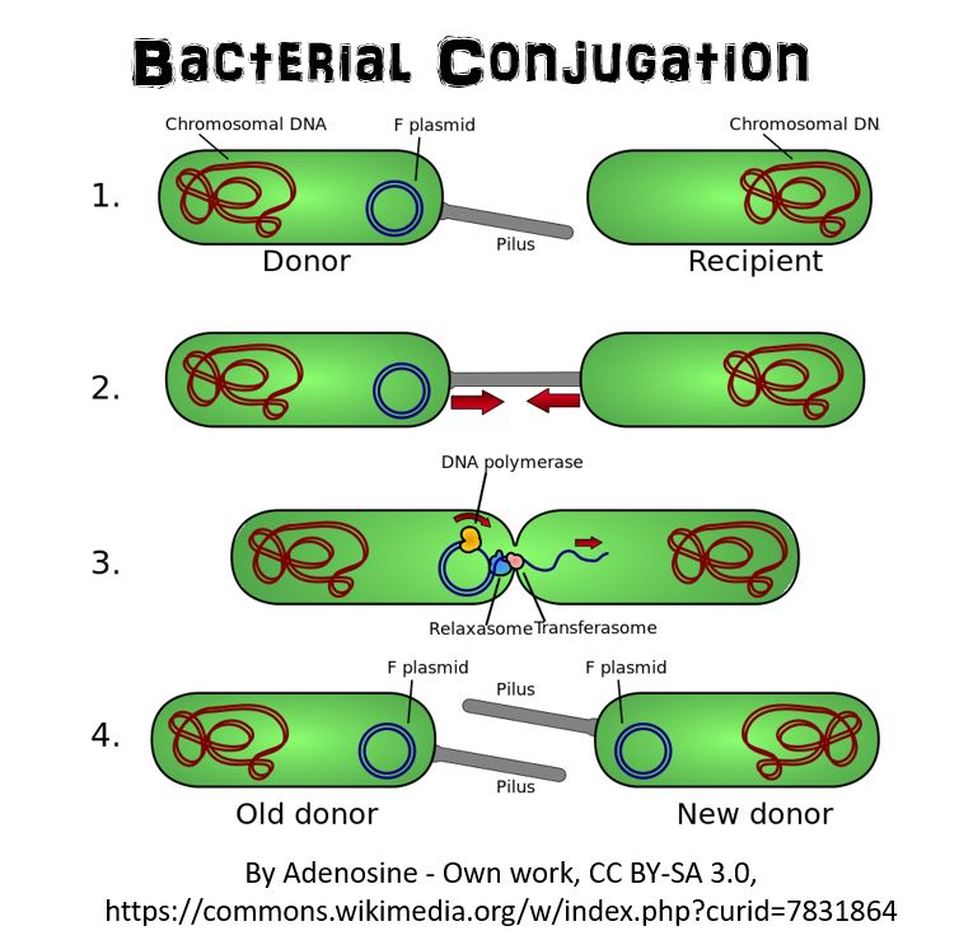
2) BACTERIAL CONJUGATION
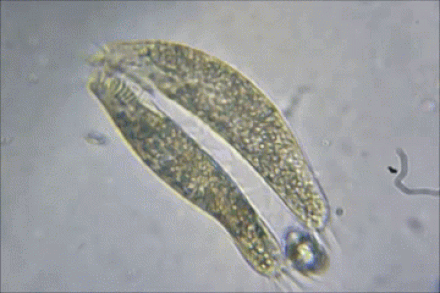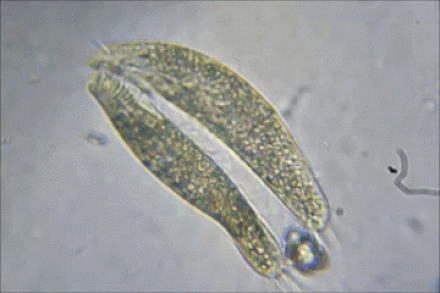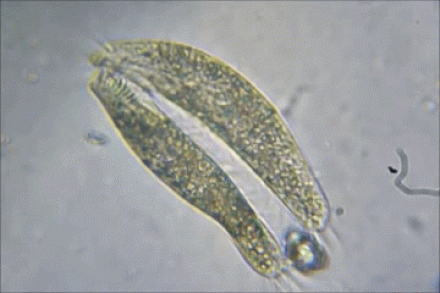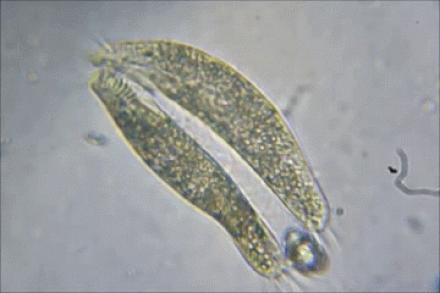 Bacterial Conjugation
Bacterial Conjugation
In bacterial conjugation, bacterial cells are able to exchange their genetic material! This is kind of like sex for bacteria! This is done using what is called the "F plasmid". The F plasmid is the "fertility plasmid". It contains genes that allow bacteria to form a sex pilus (pili is plural) which is a protrusion that can penetrate the cell wall and membrane of another bacterium. The sex pilus allows the bacteria to form a cytoplasmic bridge with another bacteria. Once that cytoplasmic bridge forms, that plasmid can then be replicated and the copy can be transferred to the other bacterium. When the bacteria receives its copy of the F plasmid, it then has the ability to now create its own sex pili and can go give copies of its F plasmid to other bacteria.
Gene Expression
THE CENTRAL DOGMA OF MOLECULAR BIOLOGY
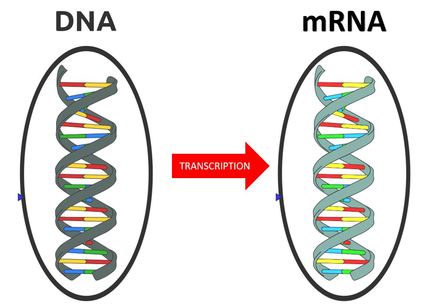
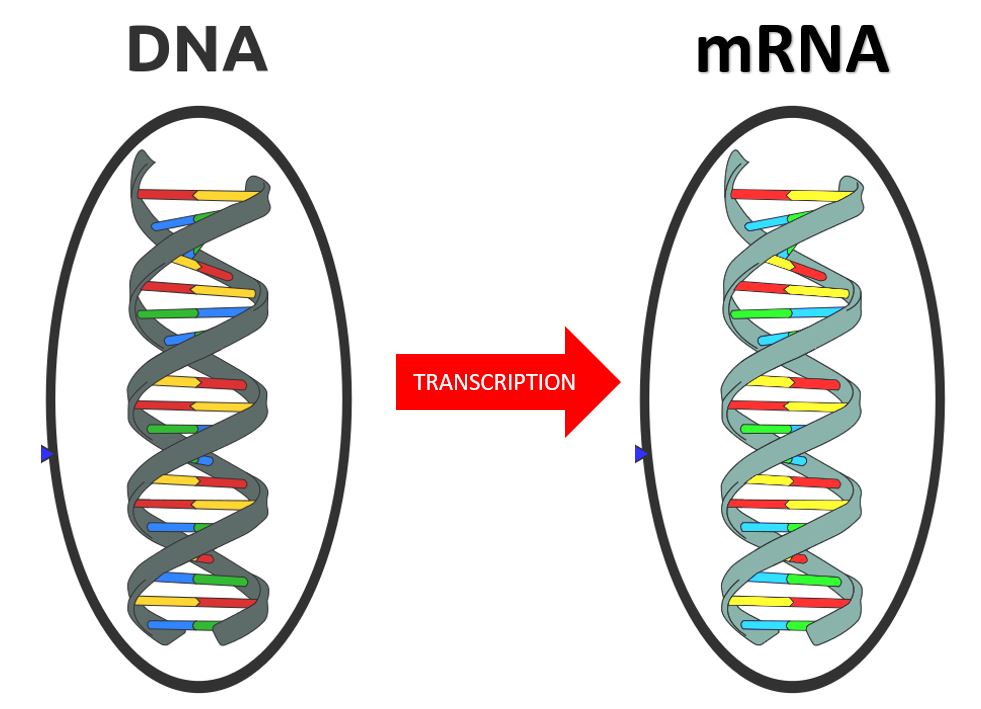
The central dogma of molecular biology explains how the information encoded in DNA sequences (genes) is expressed as proteins in a two-step process:
Step One) Transcription
Step Two) Translation
Transcription involves making mRNA using DNA as a template. Translation then translates the mRNA sequence into an amino acid sequence to form a protein.
Step One) Transcription
Step Two) Translation
Transcription involves making mRNA using DNA as a template. Translation then translates the mRNA sequence into an amino acid sequence to form a protein.
|
The genome is organized into specific sequences of nucleotides that lead to specific characteristics or features called genes. These genes cause such characteristics or features to come about through the process of transcription followed by translation. These genes are transcribed as messenger RNA (mRNA), which then gets translated into individual amino acids that will be linked together like beads on a strings to make a protein. In other words, the genome specifies the amino acid sequence that makes up the protein.
|
The genotype of a cell is the actual genes contained in a cell. Only some of the genes that make up the cell's genotype gets expressed (turned into protein through transcription and translation). For this reason, we make a distinction between the genotype itself, which is the actual genes of the cell, and the phenotype, which refers to the characteristics of a cell that are a product of the expression of only some of those genes. full collection of genes a cell contains. The phenotype is a cell’s observable characteristics resulting from the expression of specific genes proteins it is producing at a given time under specific environmental conditions.
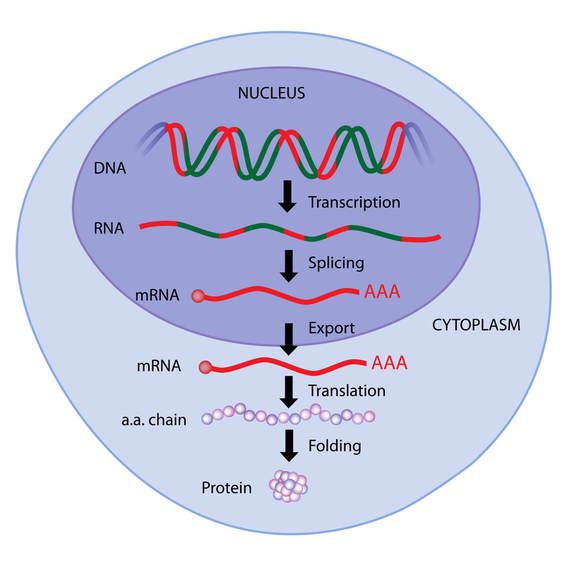
Protein synthesis in bacteria cells is much simpler than it is in eukaryotic cells. Bacterial DNA does not contain introns (regions of DNA that do not code for protein). Also, the bacterial has no nucleus for transcription to take place in when the DNA gets transcribed into messenger RNA. The process in bacteria cells, does not involve any processing of the messenger RNA. The messenger RNA will simply travel directly to a ribosome, where it will then be translated into a protein.
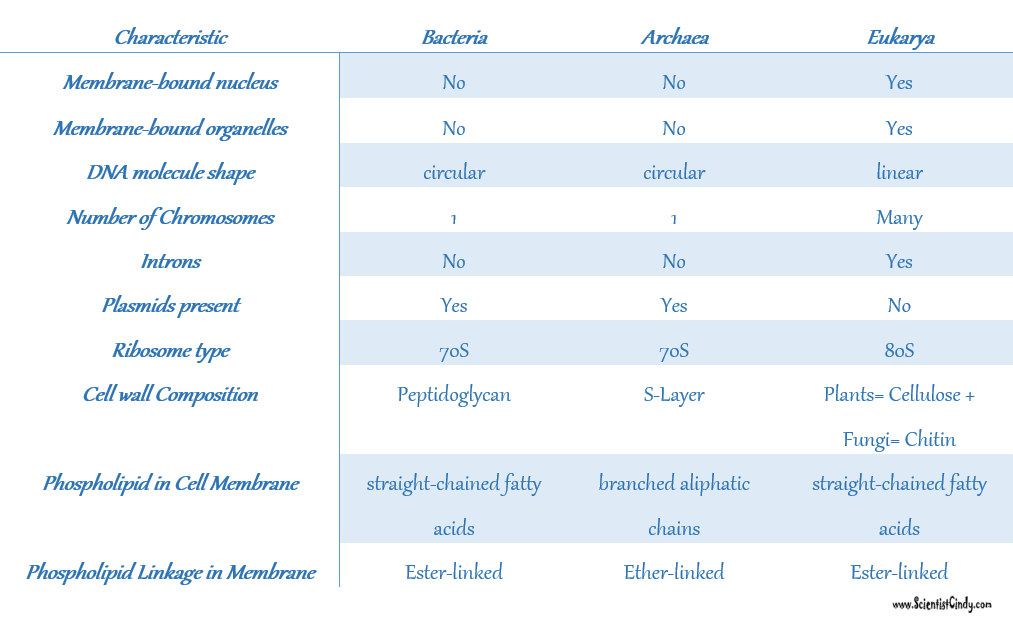
Interestingly, ARCHAEA DO HAVE INTRONS.
Interestingly, ARCHAEA DO HAVE INTRONS.
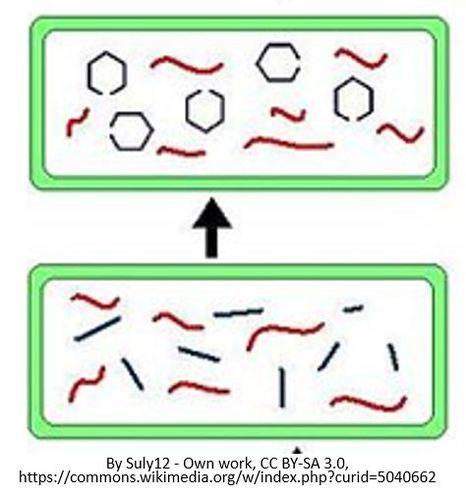
RNA TRANSCRIPTION
RNA transcription in bacteria is done in the cytoplasm, not in the nucleus like in eukaryotic cells. Transcription in bacteria is done using an enzyme called RNA polymerase. RNA Polymerase functions in a similar manner as DNA Pol III does in DNA Replication in Bacteria. RNA Polymerase adds nucleotides according to their complementary bases using a free 3’-OH group of the growing nucleotide chain.
INITIATION (of RNA Transcription)
RNA transcription requires a promoter. A promoter is a DNA sequence that transcription factors bind to. Promoters are located “upstream” from the sequences that code for proteins. The nucleotides that code for proteins would be considered to be located “downstream” from the initiation site (which is where transcription begins).
Promoters regulate transcription. They decide what gets transcribed and when it gets transcribed, if ever. Promoters are usually located between –10 and –35 bases upstream from the initiation site (designated +1). All species of bacteria have two nucleotide sequences that have been conserved, known as "consensus sequences". The 2 consensus sequences are as follows:
Promoters regulate transcription. They decide what gets transcribed and when it gets transcribed, if ever. Promoters are usually located between –10 and –35 bases upstream from the initiation site (designated +1). All species of bacteria have two nucleotide sequences that have been conserved, known as "consensus sequences". The 2 consensus sequences are as follows:
- The –10 consensus sequence, called the TATA box, is TATAAT.
- The –35 sequence is recognized and bound by Sigma.
ELONGATION (of RNA Transcription)
|
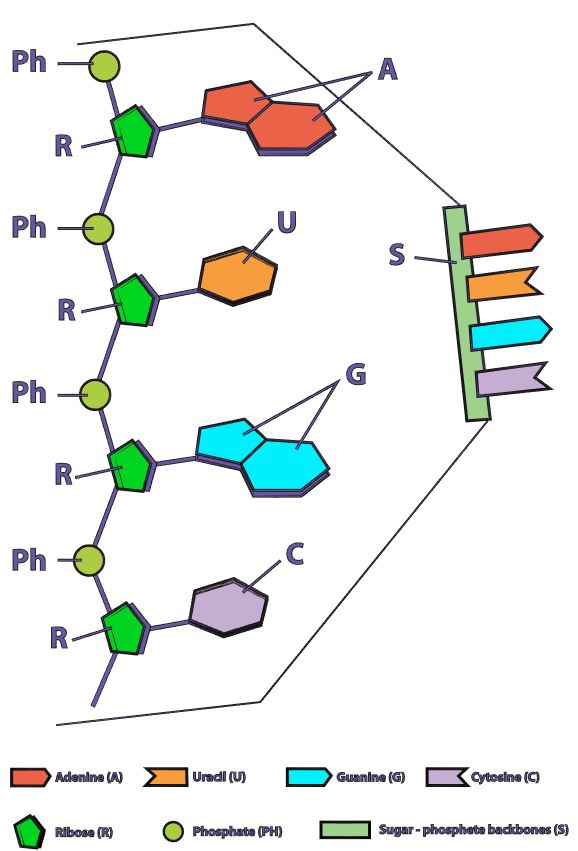
Once all of the relevant transcription factors have bound to the promoter, the elongation in transcription phase begins. In this part of RNA transcription, the sigma subunit of the RNA polymerase falls off which allows RNA transcription to begin. The RNA nucleotides are added in a complementary fashion to the DNA template in a 5’ to 3’ direction at a rate of approximately 40 nucleotides per second. RNA does not have thymine (T), so when the DNA template contains a nucleotide with an adenine (A) base, it will add a nucleotide with Uracil (U) instead.
|
Termination (of RNA Transcription)
Transcription of RNA is terminated when the bacterial RNA polymerase dissociates from the DNA template and releases the newly made RNA. This is accomplished through a terminating sequence (a repeated nucleotide sequence) that signals the RNA polymerase to terminate the process.
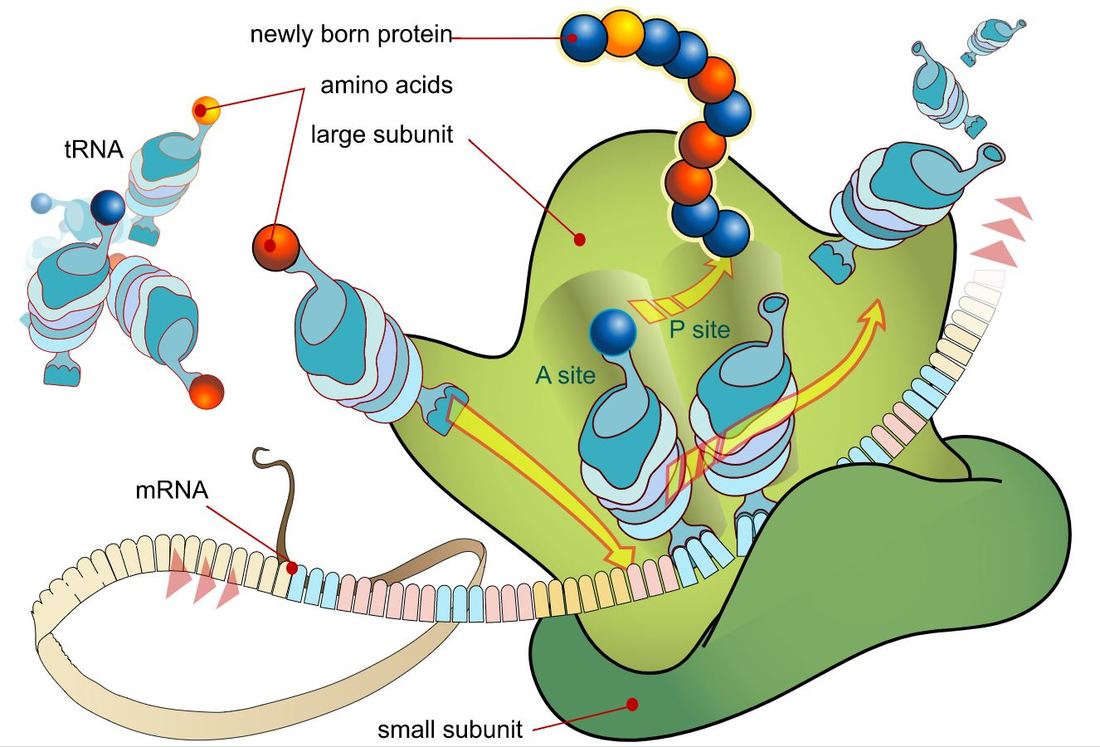
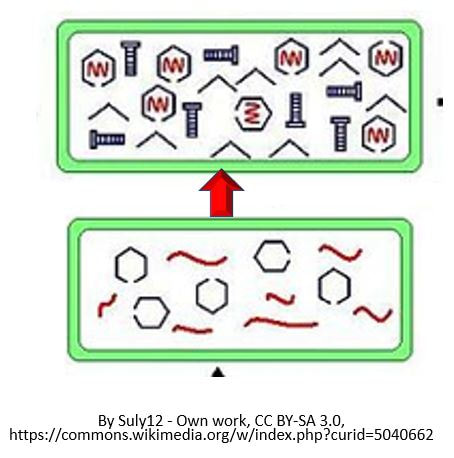
TRANSLATION
The Process of Translation of mRNA to Protein Involves the Following 3 Steps:
Genes are segments of nucleic acid sequences that will code for proteins that exhibit a specific genetic trait. The process by which the genetic sequence gets expressed into protein is a two-step process. The first step is transcription which is explain above. In short, transcription involves using the DNA molecules as a template to form messenger RNA (mRNA). The second and final step of this process is called translation. Translation is the process by which the mRNA sequence will be translated into an amino acid sequence which makes up a protein.
- Initiation
- Elongation
- Termination Steps
Genes are segments of nucleic acid sequences that will code for proteins that exhibit a specific genetic trait. The process by which the genetic sequence gets expressed into protein is a two-step process. The first step is transcription which is explain above. In short, transcription involves using the DNA molecules as a template to form messenger RNA (mRNA). The second and final step of this process is called translation. Translation is the process by which the mRNA sequence will be translated into an amino acid sequence which makes up a protein.
INITIATION (of Translation)
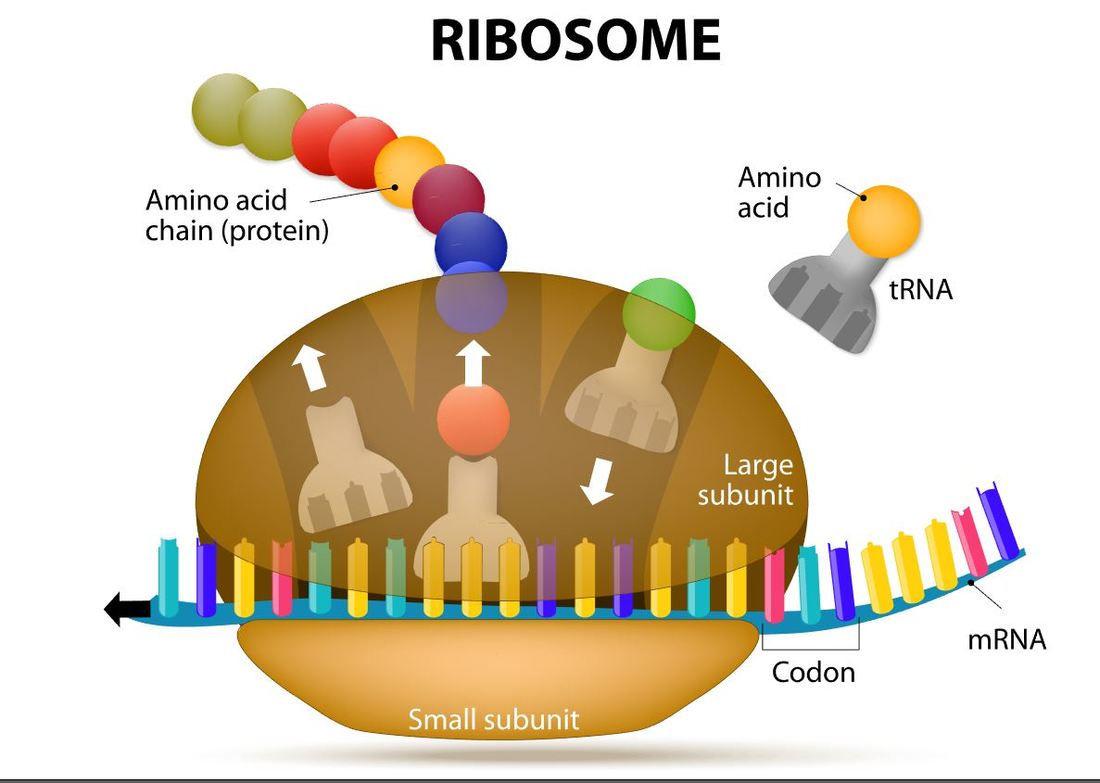
Initiation In bacteria, translation of mRNA into an amino acid sequence begins with the formation of an initiation complex. The initiation complex is formed when the mRNA, the tRNA, and the first amino acid all come together within the ribosome. The nucleotide sequence of mRNA is read in triplets called codons. All translation begins with what is called "the start codon" which is AUG. This means that the first 3 bases on the mRNA are Adenine (A), Uracil (U) and Guanine (G) = AUG. The AUG codon codes for the amino acid methionine.
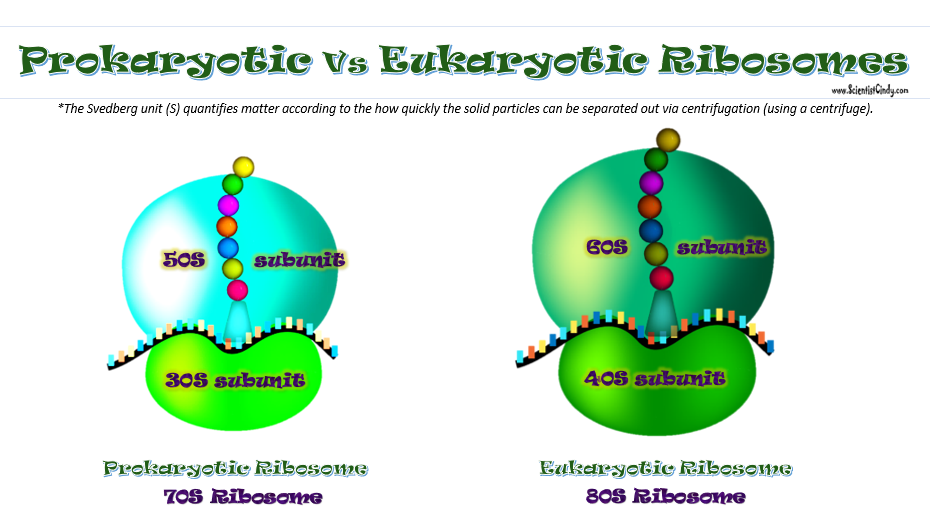
The types of ribosomes found in bacteria and archaeal cells are different from one another. The subunits differ in size, but the function remains the same. Ribosomes function as the site of protein synthesis in both eukaryotic and prokaryotic cells. Prokaryotic cells do not have membrane-bound organelles. This means that prokaryotic organisms will not have a rough endoplasmic reticulum that holds some of the ribosomes, like we see in eukaryotic cells.
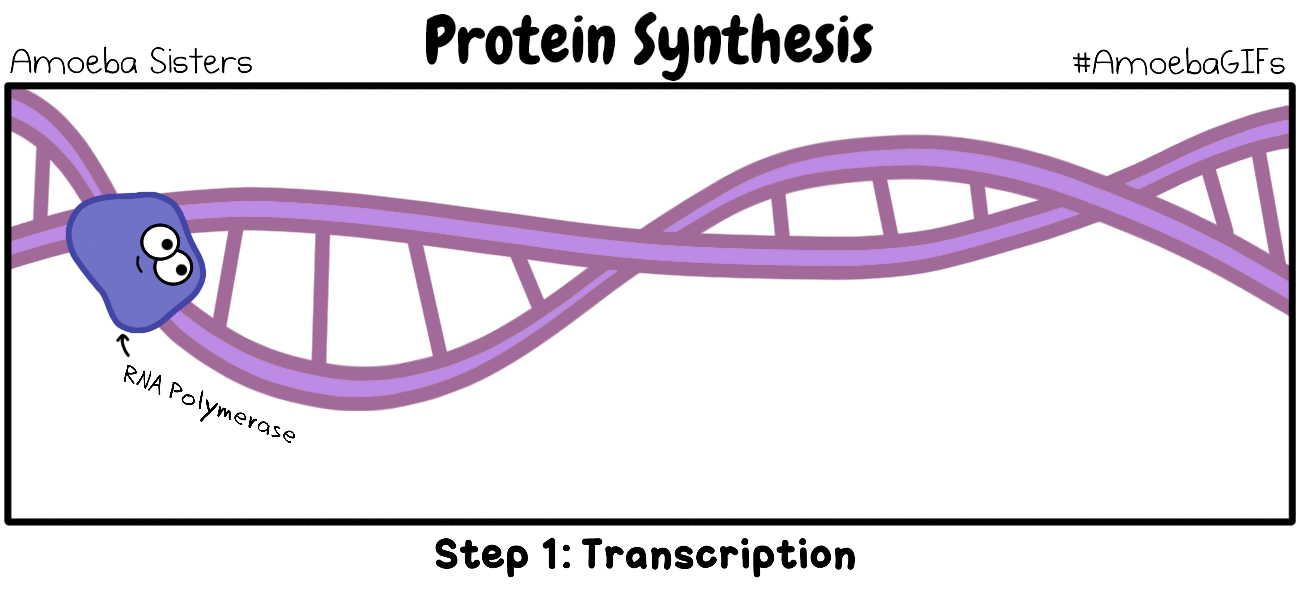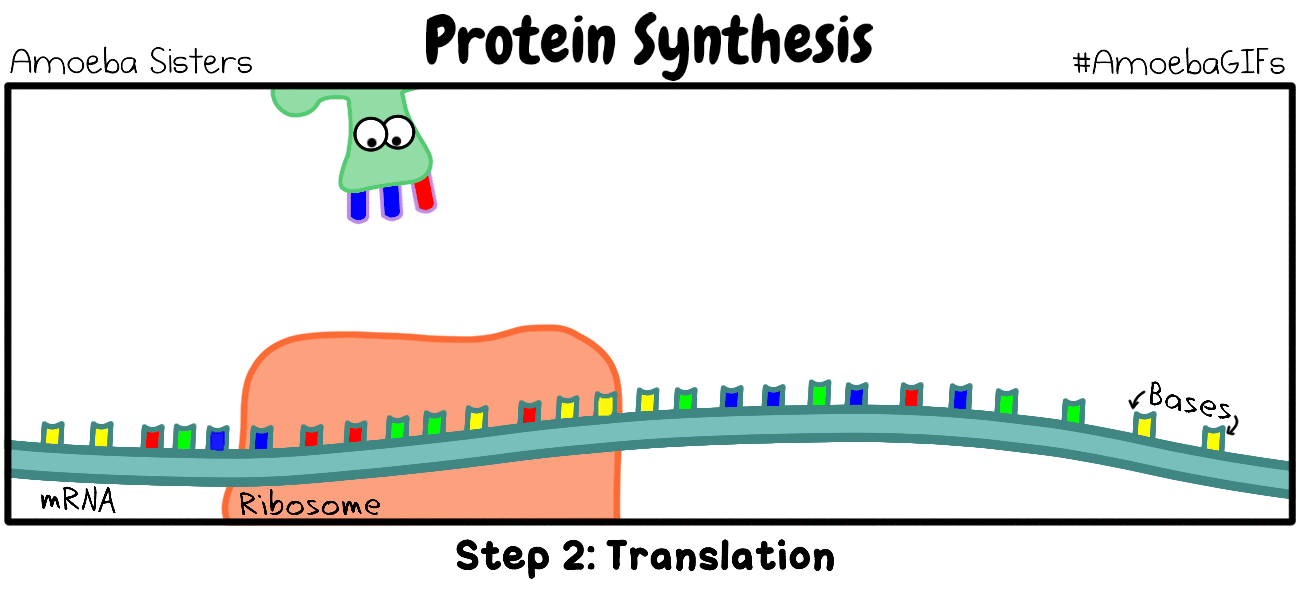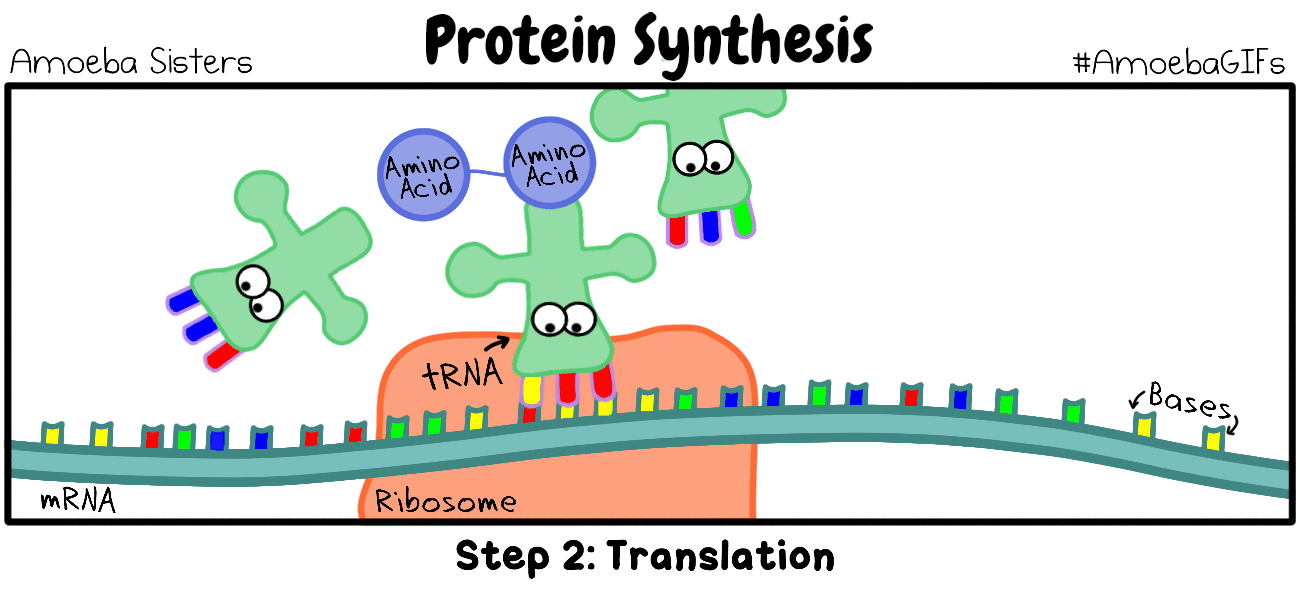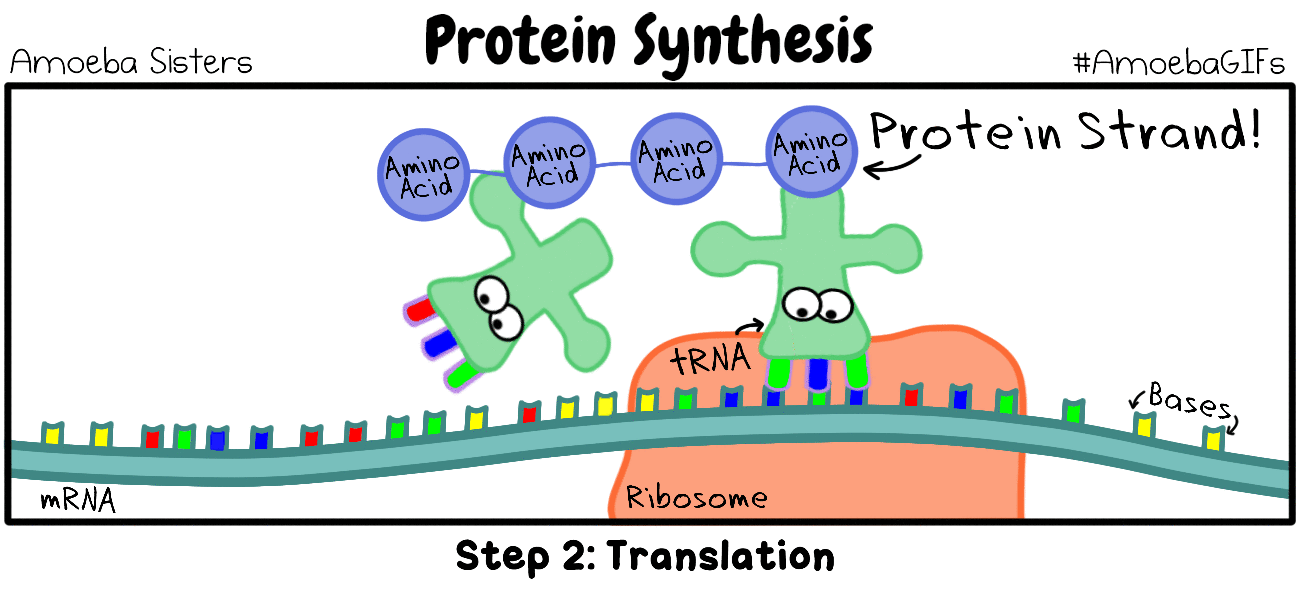
Elongation (of translation)
Elongation is the process by which amino acids are added to the amino acid sequences via peptide bonds. The amino acid chain will continue to grow and will shift in position within the ribosome.
|
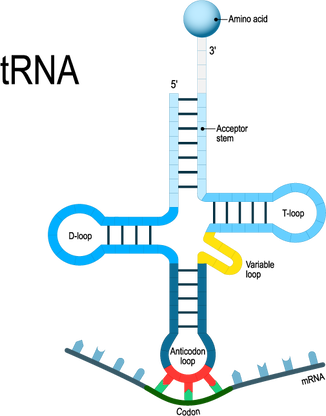
Transfer RNA will translate the mRNA code one codon at a time. For each codon, the tRNA will make an anticodon consisting of the complementary base (A pairs with U and C pairs with G). So the start codon is AUG. The tRNA (transfer RNA) will create and anticodon using complementary base-pairing. The AUG codon will result in the UAC anticodon on the tRNA. The tRNA will then bring the methionine amino acid to the ribosome as the first amino acid in the amino acid sequence which will become a protein. |
Termination of protein synthesis occurs at the stop codon. There are three different codons which are considered stop codons (UAA, UAG and UGA). When one of these stop codons reaches the ribosome, the last amino acid will break free from the ribosome and from the tRNA. The protein is now complete.
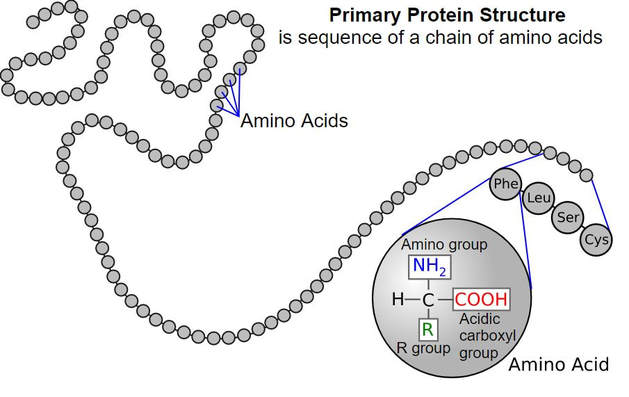
Remember that proteins are molecules that are essential for metabolism and are used to build structures within the cell. Proteins are composed of a long string (a polymer) of individual units called amino acids. The amino acids are joined together like beads on a string to make the protein.
Remember that proteins are molecules that are essential for metabolism and are used to build structures within the cell. Proteins are composed of a long string (a polymer) of individual units called amino acids. The amino acids are joined together like beads on a string to make the protein.
GENETICS of VIRUSES
Viral Genetics
|
The virus uses the host cell's machinery in order to reproduce itself. Every virus has a specific "host range". The host range is the particular type of cell or cells that the virus will be able to infect. For example, the HIV virus will only infects the helper t-cells. The helper T-cells are a specific type of white blood cells of the immune system
|
Eukaryotic cells include animal cells, like the one shown in the illustration here. Eukaryotic cells have a nucleus and have membrane bound organelles.
Viruses are considered to be a "true parasite". A parasite is an organism that depends completely on a host organisms for its survival and reproductive processes, and does so at a cost to the host. Viruses are completely dependent upon a host for their survival because they do not have any enzymes or ribosomes.
|
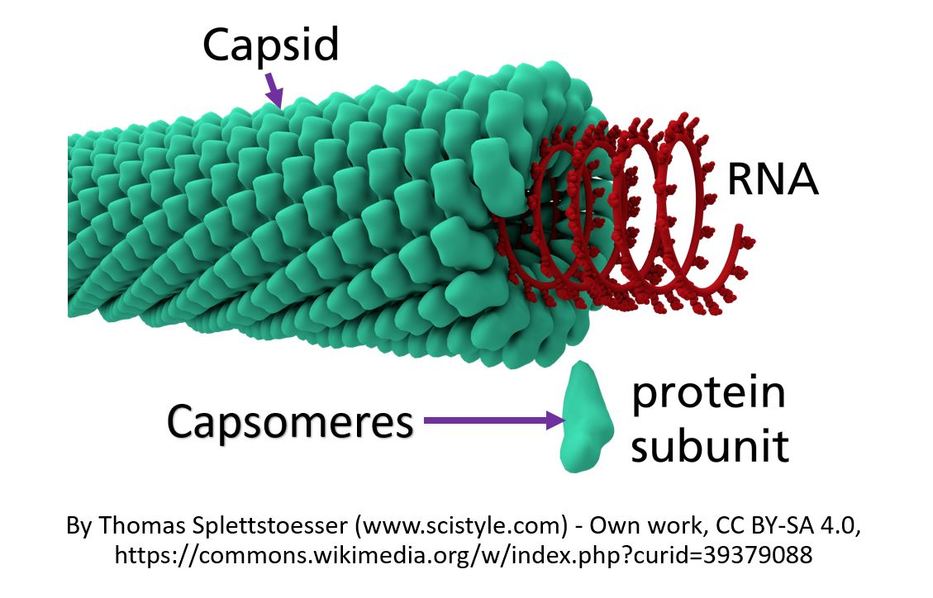
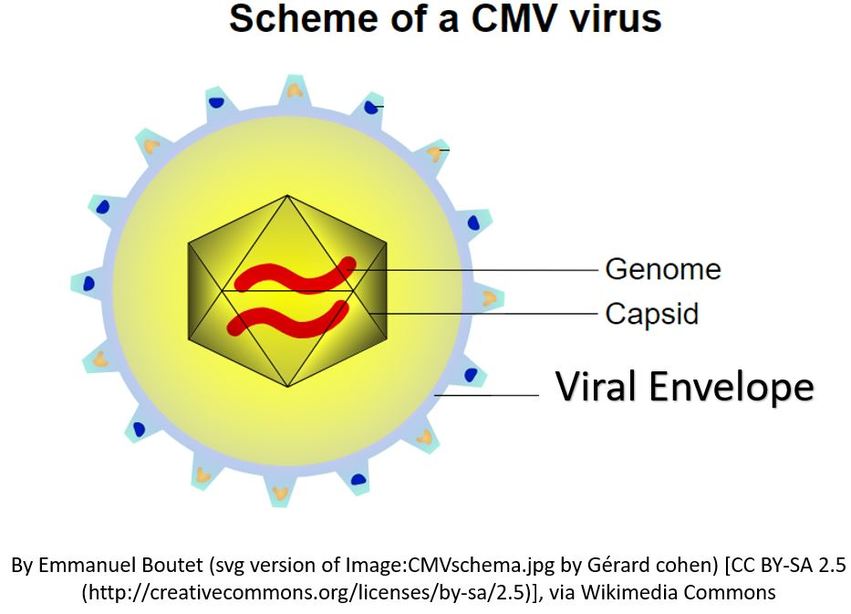
Viruses are much smaller than bacteria. Each virus is made up of genetic information which can be either RNA or DNA. The genetic material is surrounded by a protein coat that is called the capsid.
The capsid is made up of proteins called capsomeres. The capsomere is a subunit of the capsid, an outer covering of protein that protects the genetic material of a virus. Capsomeres self-assemble to form the capsid.
The capsid is made up of proteins called capsomeres. The capsomere is a subunit of the capsid, an outer covering of protein that protects the genetic material of a virus. Capsomeres self-assemble to form the capsid.
In addition to the capsid, some viruses also have another structure called the viral envelope or coat. The viral envelope consists of a lipid bilayer derived from a host cell. In addition to the lipids, the viral envelope will also contain elements of the host cell's membrane that gives the virus some type of survival advantage. For example, the viral envelope may give the virus access to specific cells or may assist the virus in evading the host's immune system. The viral envelope can contain regions of membrane that allows the virus to enter the host cell via endocytosis and to leave the host cell via exocytosis.
The life cycle of a virus depends upon the successful invasion of a host cell. Let's take a look at how viral reproduction takes place. A virus is an intracellular parasite that can reproduce only by taking over a host cell. A virus is composed of an nucleic acid genome (RNA or DNA) enclosed in a protein shell called the capsid. Some viruses are covered by a viral envelope in addition to the capsid. The viral envelope is made from the membrane of the host cell.
|
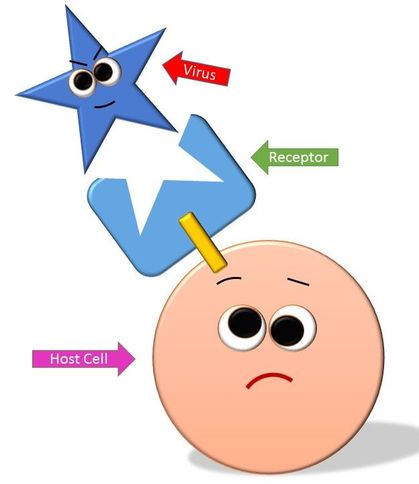
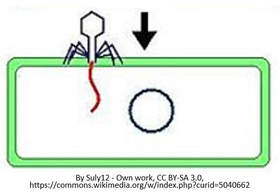
There are many strategies viruses use to attach to the host cell, but usually this process is done by a lock-and-key mechanism that will occur between the proteins of the capsid on the virus and the receptors of the host cell. In this way, the virus attaches itself to the host cell and inserts its viral DNA (or RNA) into the host cell.
Once the viral DNA/RNA enters the host cell, the viral DNA/RNA will use the host cell's nucleotides and enzymes to replicate itself making more copies of the viral genome.. |
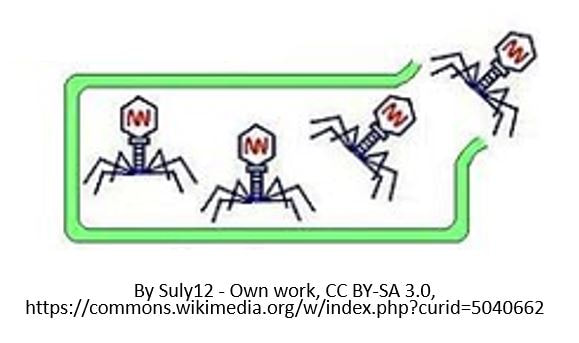
The viral DNA/RNA then uses the host cell's materials and machinery to transcribe its genes into messenger RNA (mRNA). The mRNA template is then used to translate message into an amino acid sequence that makes up the viral protein. Once all of the necessary building blocks have formed, the viral proteins produced, the viral DNA and capsid proteins will self-assemble into new viruses. The new viruses exit the host cell, often destroying the host cell in the process. The new viruses can go on to infect other cells, thereby spreading the viral infection.
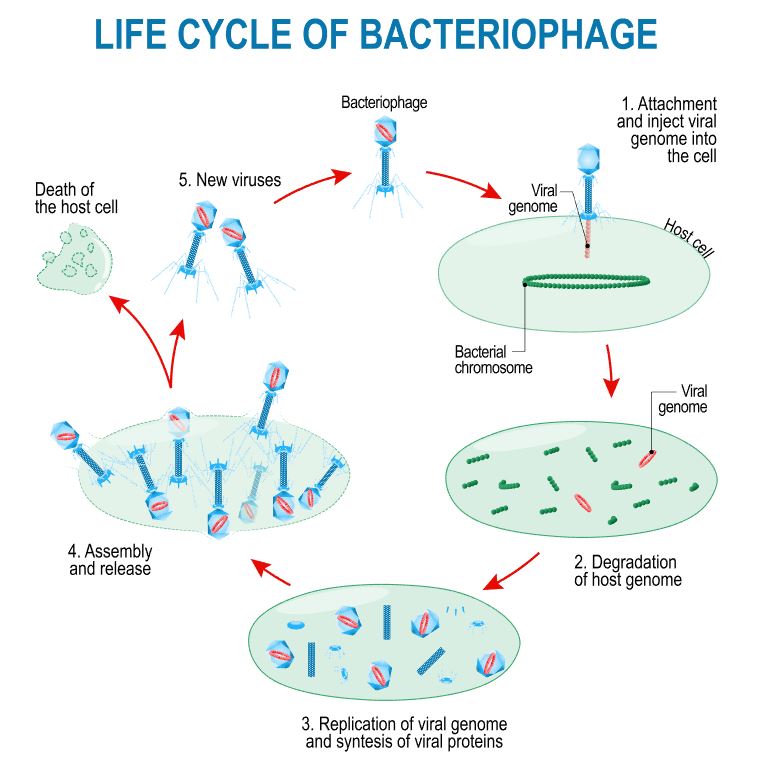
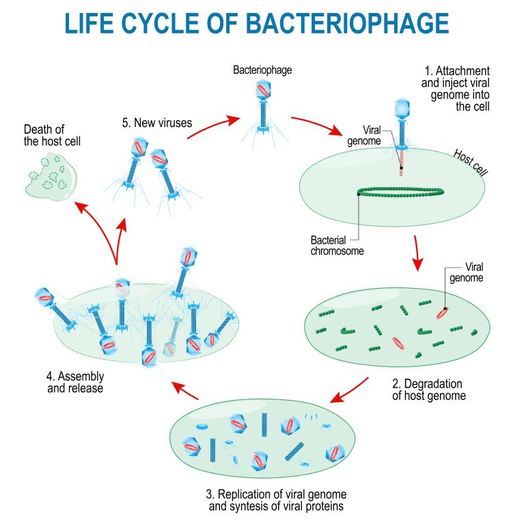
The Lytic Cycle and the Lysogenic Cycle
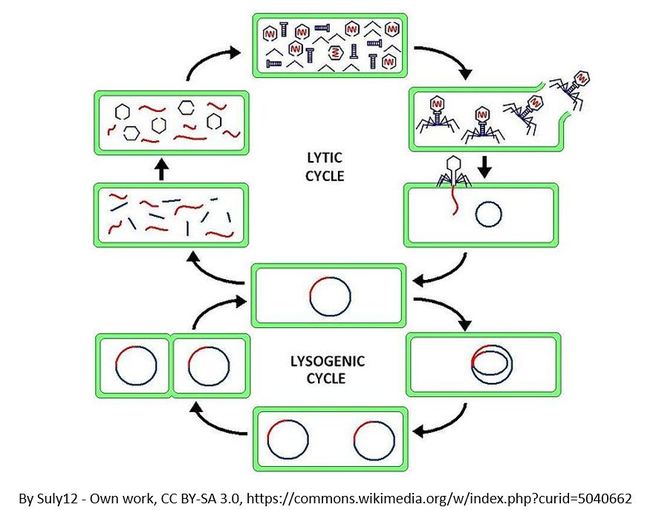
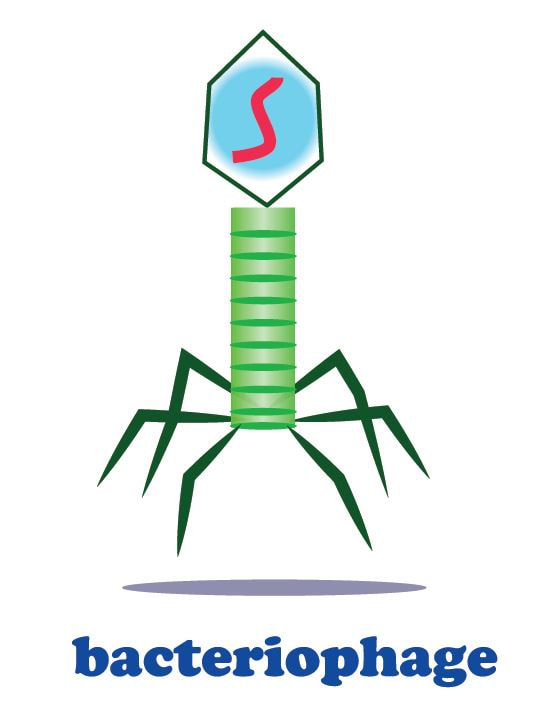
Bacteriophages are viruses that infect bacteria cells. There are two types of reproductive cycles that a bacteriophage can enter; 1) the lytic cycle and 2) the
lysogenic cycle..
The lytic cycle results in lysis of the host cell, which ends in death. In the lysogenic cycle, the viral genome enters the nucleus and inserts itself into the host genome, keeping the host cell alive.
lysogenic cycle..
The lytic cycle results in lysis of the host cell, which ends in death. In the lysogenic cycle, the viral genome enters the nucleus and inserts itself into the host genome, keeping the host cell alive.
Let's look at an overview of the lytic cycle.
|
|
Let's look at an overview of the lysogenic cycle.
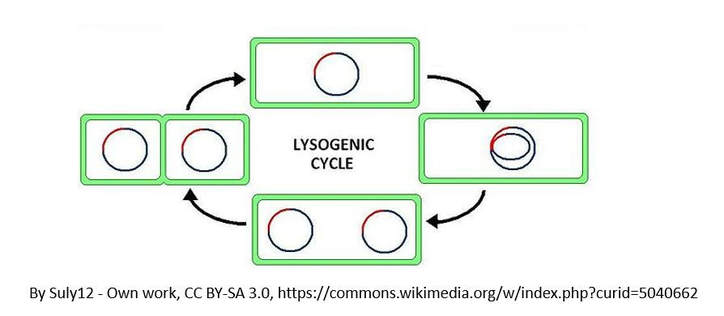
The other reproductive cycle that a phage virus can use is called the lysogenic cycle. Unlike the lytic cycle, the lysogenic cycle is not going to kill the host cell. The viruses that reproduce
using this method are considered "temperate" viruses.
using this method are considered "temperate" viruses.
- First, the phage infects the host bacteria.
- Then, the viral DNA (or RNA) will be inserted into the bacteria host.
- The Viral DNA is then going to incorporate itself into the host cells genome.
- Once the virus's DNA has been incorporated into the host cells DNA, it is now considered a prophage.
- Each time the infected bacteria cell reproduces itself, it will also reproduce the viral DNA
- After some time has past, the prophage will exit the host cell's genome and the virus will begin to undergo the deadly lytic cycle.
THE LIFE CYCLE OF RETROVIRUSES
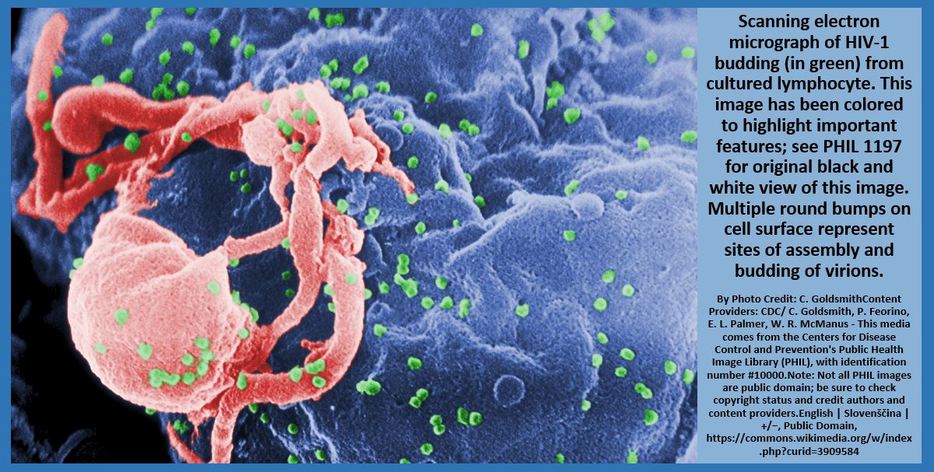
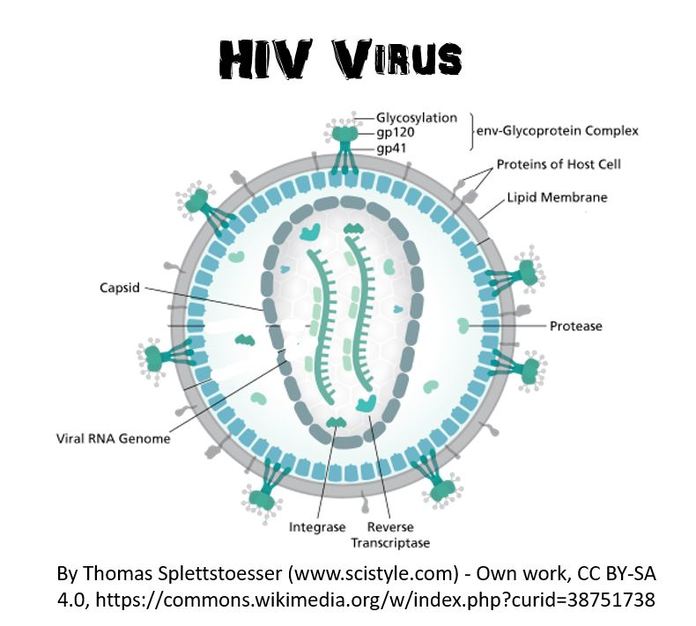
Some viruses are classified as retroviruses due to the way in which they reproduce their genetic material in the host cell. Arguably, the best known retrovirus is the human immunodeficiency virus (HIV).
The HIV virus is called a retrovirus due to the way that it reproduces in the host cell. HIV virus infects white blood cells.
|
The HIV virus has a viral envelope that surrounds the capsid.
The viral envelope is composed of pieces of the host cell's membrane. The viral envelope allows the HIV virus to enter the host cell (white blood cell) through endocytosis. In addition, the viral envelope will also enable the virus to exit the white blood cell through exocytosis. |
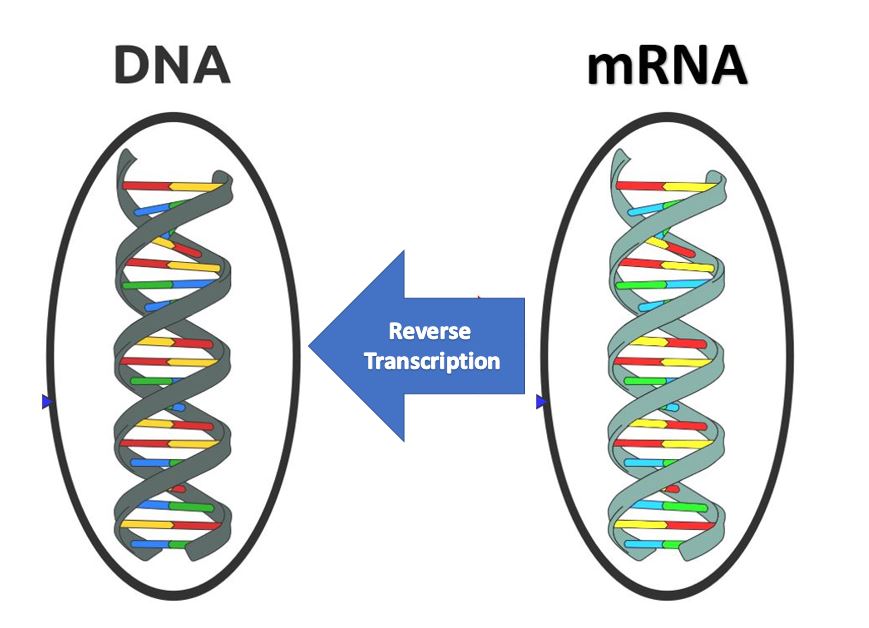
The HIV virus is considered a retrovirus, because it has RNA as its genetic information instead of DNA. The HIV virus has a capsid (or protein coat) and 2 strands of single-stranded RNA. In addition to the single-stranded RNA, the HIV virus contains a special enzyme called reverse transcriptase. Reverse transcriptase is an enzyme that is able to essentially perform transcription in reverse!
Recall that in normal transcription, the DNA is replicated as mRNA. Now in a retrovirus, the viral single-stranded RNA will use reverse transcriptase to build a double-stranded DNA molecule from the RNA, using the nucleotide sequence of the RNA as a template.
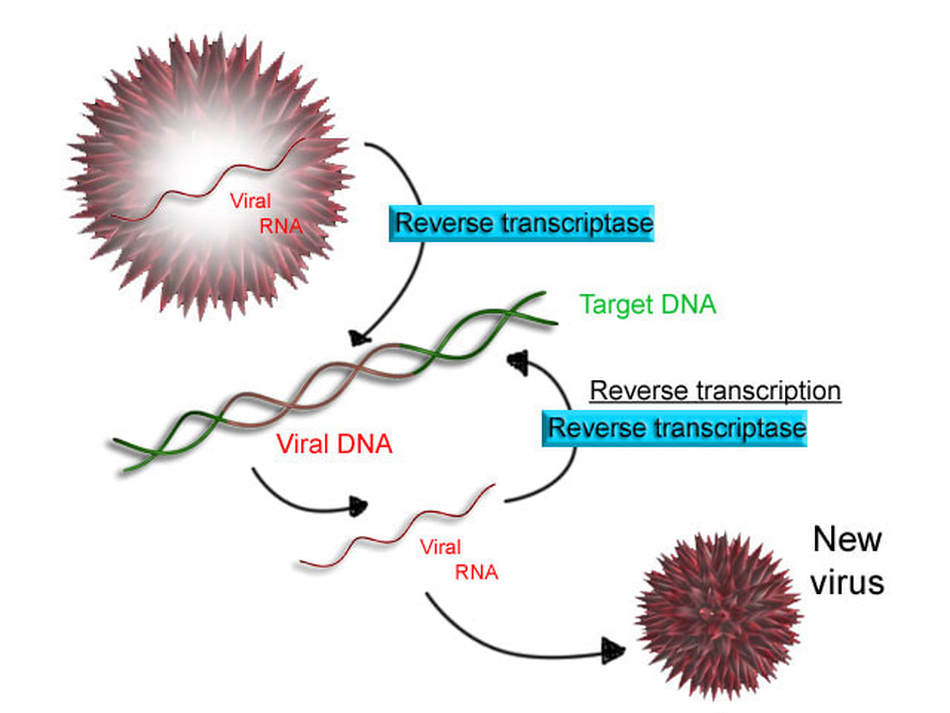
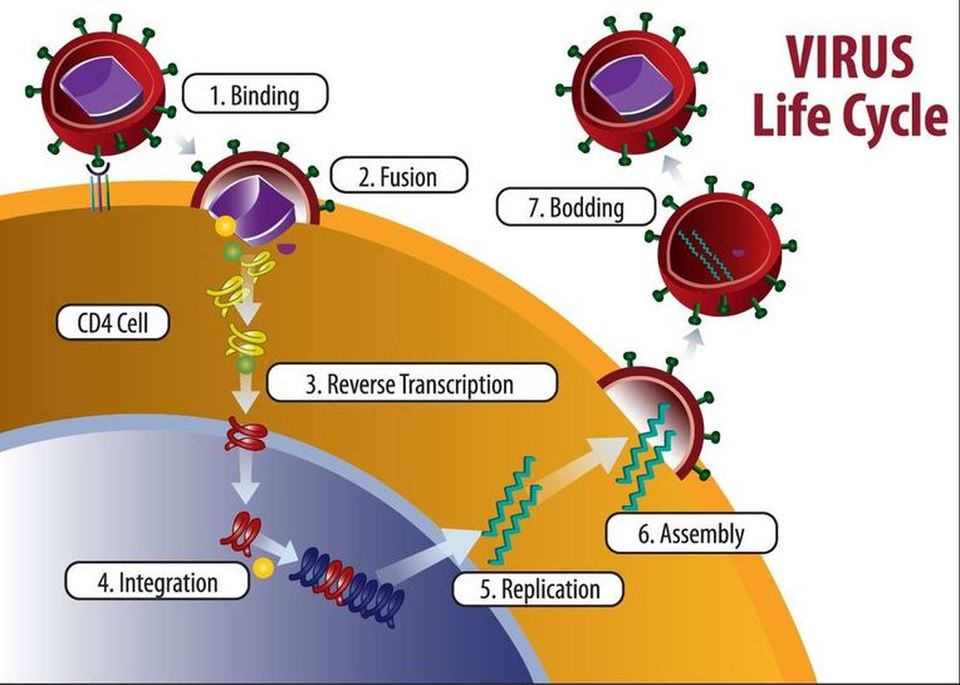
THE RETROVIRUS LIFE CYCLE
- HIV infects a class of white blood cells called helper t-cell
- The HIV virus enters the host cell through endocytosis
- Once the viral RNA has entered the host cell, reverse transcriptase is going to "reverse transcribe" the viral RNA into DNA.
- After the viral DNA has been created, it will incorporate itself into the DNA of the host cell (the helper t-cell). At this point, this is called a provirus.
- A provirus is the same thing as a pro phage, except that pro phages are found in bacteria where pro viruses are found in eukaryotic cells
- Every time the white blood cell goes through protein synthesis, new viral RNA is generated.
- The viral RNA that is generated will eventually be used to assemble new HIV viruses inside the white blood cell.
- The viruses will exit the white blood cell through exocytosis.
- When exocytosis takes place, the viral envelope will be formed,
- When these viruses exit the host cell the host cell is going to be destroyed as an end result of this life cycle.